高斯混合体的数学概念
高斯混合体是由若干个高斯随机变量线性组合而成,表示为
GMM
=
∑
k
=
1
K
α
k
?
G
a
u
s
s
i
a
n
k
?
,
?
∑
k
α
k
=
1
?
,
?
α
k
>
0
?
,
?
?
α
k
\text{GMM} = \sum_{k=1}^K \alpha_k \cdot Gaussian_k\ ,\ \sum_k \alpha_k =1\ ,\ \alpha_k>0\ ,\ \forall \alpha_k
GMM=k=1∑K?αk??Gaussiank??,?k∑?αk?=1?,?αk?>0?,??αk?
有
K
K
K个分量,也称簇。该模型主要面向聚类任务,基本观点为“数据从某一簇生成”。对于新输入的数据,需要模型给出属于哪一个簇的判断。
生成GMM分布模型
需要指定的参数有:簇的数量
K
K
K,各簇的均值(位置)
μ
i
\mu_i
μi?、协方差矩阵(因为是二维)以及混合比例向量
α
\alpha
α。考虑到限制条件
Σ
k
α
k
=
1
?
,
?
α
k
>
0
?
,
?
?
α
k
\Sigma_k \alpha_k=1 \ ,\ \alpha_k>0\ ,\ \forall \alpha_k
Σk?αk?=1?,?αk?>0?,??αk?,选择从狄利克雷分布np.random.dirichlet生成
α
\alpha
α,满足限制条件。出于简单实验的目的,直接指定了
μ
?
,
?
c
o
v
\mu\ ,\ cov
μ?,?cov,实际上也可以随机指定,满足各分量协方差矩阵正定即可。
检验模型
将模型生成的数据与解析数据放在一起可视化。
代码
import math
import numpy as np
import pandas as pd
import seaborn as sns
import matplotlib as mpl
import matplotlib.pyplot as plt
from sklearn import mixture
from matplotlib.colors import LogNorm
from mpl_toolkits.mplot3d import Axes3D
from matplotlib import cm
%matplotlib
#样本数量
n_samples = 20000
#分量数量
K=5
#均值矩阵
mu = np.array([[0, 0],[10, 0],[5, 5],[0, 10],[10, 10]])
#协方差矩阵
cc = np.array([1,0.2,0.2,1,1,0.4,0.4,1,1,0,0,1,1,-0.6,-0.6,1,1,-0.8,-0.8,1])
cov = cc.reshape((5,2,2))
cov_inverse = np.linalg.inv(cov)#用于后续生成解析数据
#权重矩阵
alpha = np.random.dirichlet(np.array([3,4,5,6,7]))
#随机数据装载
gmm_0 = np.random.multivariate_normal(mu[0],cov[0],n_samples)
gmm_1 = np.random.multivariate_normal(mu[1],cov[1],n_samples)
gmm_2 = np.random.multivariate_normal(mu[2],cov[2],n_samples)
gmm_3 = np.random.multivariate_normal(mu[3],cov[3],n_samples)
gmm_4 = np.random.multivariate_normal(mu[4],cov[4],n_samples)
GMM = np.vstack([gmm_0,gmm_1,gmm_2,gmm_3,gmm_4])
#查看原始数据
#figure 1: 权重向量 alpha
alpha_d = np.around(alpha,decimals=2)
fig = plt.figure(1,figsize=(8,6))
ax = fig.add_subplot(111,title=r"$\alpha$ from dirichlet")
name = [r"$\alpha_1$",r"$\alpha_2$",r"$\alpha_3$",r"$\alpha_4$",r"$\alpha_5$"]
df = pd.DataFrame({"index":name,"value":alpha_d})
ax.bar(data=df,x="index",height="value",width=0.5,color=["blue","green","purple","red","orange"],edgecolor='black')
ax.set_xlabel(r"$\alpha_i$")
ax.set_ylabel("values")
for idx, text in zip(name, alpha_d):
ax.text(idx, text, text, ha='center', va='bottom',fontsize=12)
#figure 2: gmm各分量、gmm合并但未拟合的平面直方图
fig = plt.figure(2,figsize=(15,12))
grid_size=int(n_samples/400)
ax = fig.add_subplot(231,aspect='equal',title=r"$gmm_1$")
ax.set_xlabel('x')
ax.set_ylabel('y')
ax.hexbin(x=gmm_0[:,0],y=gmm_0[:,1],gridsize=grid_size,cmap='viridis')
ax = fig.add_subplot(232,aspect='equal',title=r"$gmm_2$")
ax.set_xlabel('x')
ax.set_ylabel('y')
ax.hexbin(x=gmm_1[:,0],y=gmm_1[:,1],gridsize=grid_size,cmap='viridis')
ax = fig.add_subplot(233,aspect='equal',title=r"$gmm_3$")
ax.set_xlabel('x')
ax.set_ylabel('y')
ax.hexbin(x=gmm_2[:,0],y=gmm_2[:,1],gridsize=grid_size,cmap='viridis')
ax = fig.add_subplot(234,aspect='equal',title=r"$gmm_4$")
ax.set_xlabel('x')
ax.set_ylabel('y')
ax.hexbin(x=gmm_3[:,0],y=gmm_3[:,1],gridsize=grid_size,cmap='viridis')
ax = fig.add_subplot(235,aspect='equal',title=r"$gmm_5$")
ax.set_xlabel('x')
ax.set_ylabel('y')
ax.hexbin(x=gmm_4[:,0],y=gmm_4[:,1],gridsize=grid_size,cmap='viridis')
ax = fig.add_subplot(236,aspect='equal',title="Gaussians before mixing")
ax.set_xlabel('x')
ax.set_ylabel('y')
ax.hexbin(x=GMM[:,0],y=GMM[:,1],gridsize=grid_size,cmap='viridis')
#加载模型,拟合
#K个分量
#covariance_type = full 每个分量具有自己的协方差矩阵
#init_paramas ='kmeans' 使用 kmeans方法拟合
#weights_init = alpha 指定权重矩阵
#means_init = mu 指定各分量中心
"""
为sklearn.mixture.GaussianMixture指定众多参数是为了更精准的拟合,
并非为了检测该分类器性能,
而是在之后使用该模型生成数据服务于其他贝叶斯任务,
成为其他推理方法的参照。
"""
model = mixture.GaussianMixture(n_components=K, covariance_type='full',init_params='kmeans',weights_init=alpha,means_init=mu)
model.fit(GMM)
#查看GMM预测的负对数似然函数值 model.score_samples
x = np.linspace(-100,100,1000)
y = np.linspace(-100,100,1000)
X, Y = np.meshgrid(x, y)
XX = np.array([X.ravel(), Y.ravel()]).T
Z = -model.score_samples(XX)
Z = Z.reshape(X.shape)
#绘制等高图查看负 log-liklihood 预测值
fig = plt.figure(3,figsize=(8,6))
ax = fig.add_subplot(111,aspect='equal',title='Negative log-likelihood predicted by a GMM')
ax.set_xlabel('x')
ax.set_ylabel('y')
CS = ax.contourf(X, Y, Z,cmap='YlGnBu', levels=25)
plt.colorbar(CS, shrink=0.8, aspect=10)
sample_test, index = model.sample(10000)
fig = plt.figure(4,figsize=(20,6))
#GMM模型中各分量的均值
ax = fig.add_subplot(131,title='model means',aspect='equal')
ax.set_xlabel('x')
ax.set_ylabel('y')
ax.scatter(x=model.means_[:,0],y=model.means_[:,1],c='r')
#查看从GMM模型生成的数据 model.samples
ax = fig.add_subplot(132,title="model sample")
ax.set_xlabel('x')
ax.set_ylabel('y')
ax.hexbin(x=sample_test[:,0],y=sample_test[:,1],gridsize=grid_size,cmap='jet')
#准备解析数据
#设置边界:正负 3σ
bound = list((
mu[0,0]-3*cov[0,0,0], mu[0,0]+3*cov[0,0,0], mu[0,1]-3*cov[0,1,1], mu[0,1]+3*cov[0,1,1],
mu[1,0]-3*cov[1,0,0], mu[1,0]+3*cov[1,0,0], mu[1,1]-3*cov[1,1,1], mu[1,1]+3*cov[1,1,1],
mu[2,0]-3*cov[2,0,0], mu[2,0]+3*cov[2,0,0], mu[2,1]-3*cov[2,1,1], mu[2,1]+3*cov[2,1,1],
mu[3,0]-3*cov[3,0,0], mu[3,0]+3*cov[3,0,0], mu[3,1]-3*cov[3,1,1], mu[3,1]+3*cov[3,1,1],
mu[4,0]-3*cov[4,0,0], mu[4,0]+3*cov[4,0,0], mu[4,1]-3*cov[4,1,1], mu[4,1]+3*cov[4,1,1],
))
#步长
step=0.1
#索引
x= np.arange(min(bound),max(bound),step)
y= np.arange(min(bound),max(bound),step)
X, Y = np.meshgrid(x,y)
#生成数据
#1. K 个独立的 2 维高斯
Z_1 = 1/np.sqrt(2*math.pi*np.linalg.det(cov[0])) * np.exp(-1/2*((
X-mu[0,0])**2*cov_inverse[0,0,0]
+(X-mu[0,0])*(Y-mu[0,1])*cov_inverse[0,1,0]
+(Y-mu[0,1])*(X-mu[0,0])*cov_inverse[0,0,1]
+(Y-mu[0,1])**2*cov_inverse[0,1,1]
))
Z_2 = 1/np.sqrt(2*math.pi*np.linalg.det(cov[1])) * np.exp(-1/2*((
X-mu[1,0])**2*cov_inverse[1,0,0]
+(X-mu[1,0])*(Y-mu[1,1])*cov_inverse[1,1,0]
+(Y-mu[1,1])*(X-mu[1,0])*cov_inverse[1,0,1]
+(Y-mu[1,1])**2*cov_inverse[1,1,1]
))
Z_3 = 1/np.sqrt(2*math.pi*np.linalg.det(cov[2])) * np.exp(-1/2*((
X-mu[2,0])**2*cov_inverse[2,0,0]
+(X-mu[2,0])*(Y-mu[2,1])*cov_inverse[2,1,0]
+(Y-mu[2,1])*(X-mu[2,0])*cov_inverse[2,0,1]
+(Y-mu[2,1])**2*cov_inverse[2,1,1]
))
Z_4 = 1/np.sqrt(2*math.pi*np.linalg.det(cov[3])) * np.exp(-1/2*((
X-mu[3,0])**2*cov_inverse[3,0,0]
+(X-mu[3,0])*(Y-mu[3,1])*cov_inverse[3,1,0]
+(Y-mu[3,1])*(X-mu[3,0])*cov_inverse[3,0,1]
+(Y-mu[3,1])**2*cov_inverse[3,1,1]
))
Z_5 = 1/np.sqrt(2*math.pi*np.linalg.det(cov[4])) * np.exp(-1/2*((
X-mu[4,0])**2*cov_inverse[4,0,0]
+(X-mu[4,0])*(Y-mu[4,1])*cov_inverse[4,1,0]
+(Y-mu[4,1])*(X-mu[4,0])*cov_inverse[4,0,1]
+(Y-mu[4,1])**2*cov_inverse[4,1,1]
))
#2. 根据 alpha 做线性组合(混合)
GMM_SUM = Z_1*alpha[0] + Z_2*alpha[1] + Z_3*alpha[2] + Z_4*alpha[3] + Z_5*alpha[4]
#3. 绘图:3d表面图,坐标面上有等高线投影
ax = fig.add_subplot(133,projection='3d',title='GMM analytical 3d surface')
ax.plot_surface(X, Y, GMM_SUM, lw=2,rstride=1, cstride=1, cmap='jet', alpha=0.8)
cset = ax.contour(X,Y,GMM_SUM,zdir='x',offset=-Y.min() if Y.min()>0 else Y.min(),cmap='coolwarm')
cset = ax.contour(X,Y,GMM_SUM,zdir='y',offset=X.max() if X.max()>0 else -X.max(),cmap='coolwarm')
cset = ax.contour(X,Y,GMM_SUM,zdir='z',offset=-abs(GMM_SUM).max(),cmap='coolwarm')
ax.set_xlabel('x')
ax.set_ylabel('y')
ax.set_zlabel('z')
ax.set_xlim(X.min(),X.max())
ax.set_ylim(Y.min(),Y.max())
ax.set_zlim(-abs(GMM_SUM).max(),abs(GMM_SUM).max())
plt.show()
运行结果为若干张图:
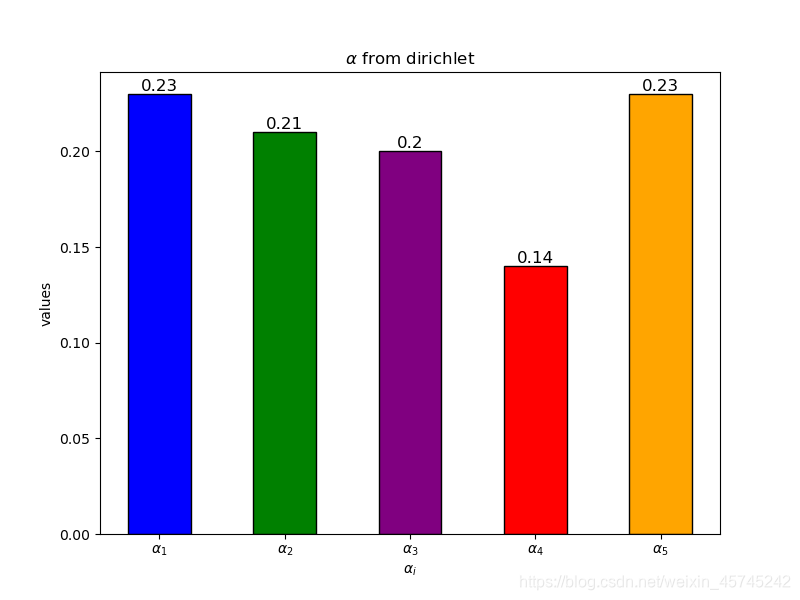


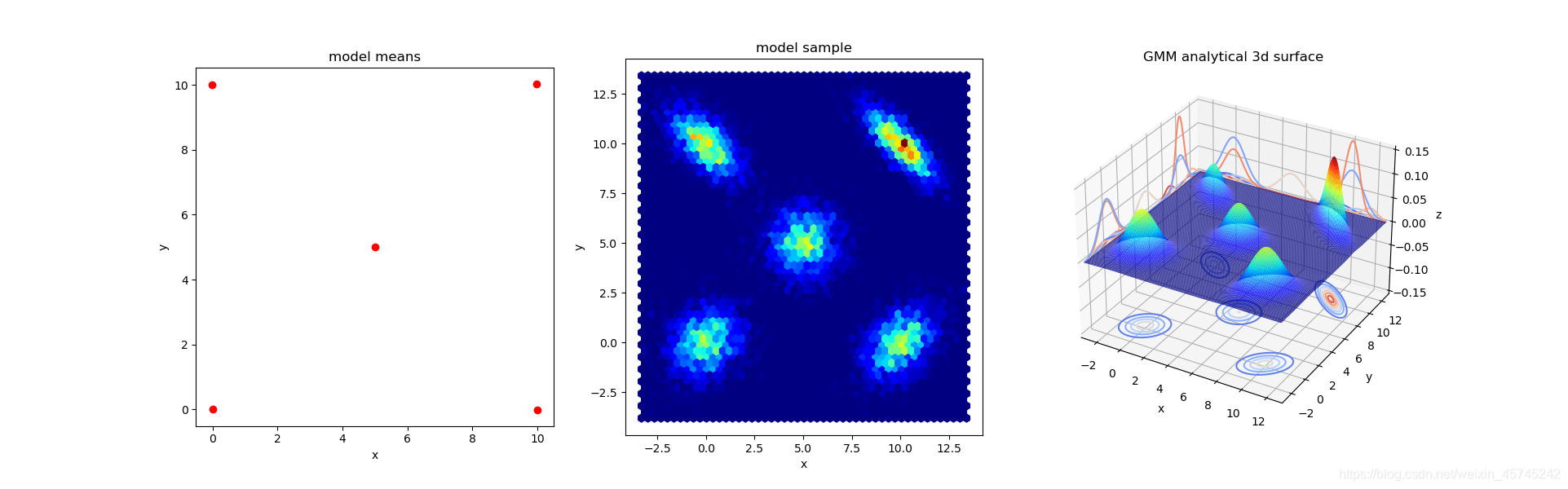
最后一张图的3d模型可通过鼠标拖动旋转查看。
最后,尝试一下模型对新数据的预测,从图像来看,各簇附近的点会被比较准确地预测到该簇。
test = model.predict([[2,2],[10,10]])
print(test)
运行结果为:
[0 4]
通过model.means_查看模型的均值确定0与4代表的簇,相符。