我目前python所有的环境基本上都是python3.6.10, 这里安装的scanpy默认是1.7.2,但是安装完,你会发现umap图出现很奇怪的现象,下面是测试的结果
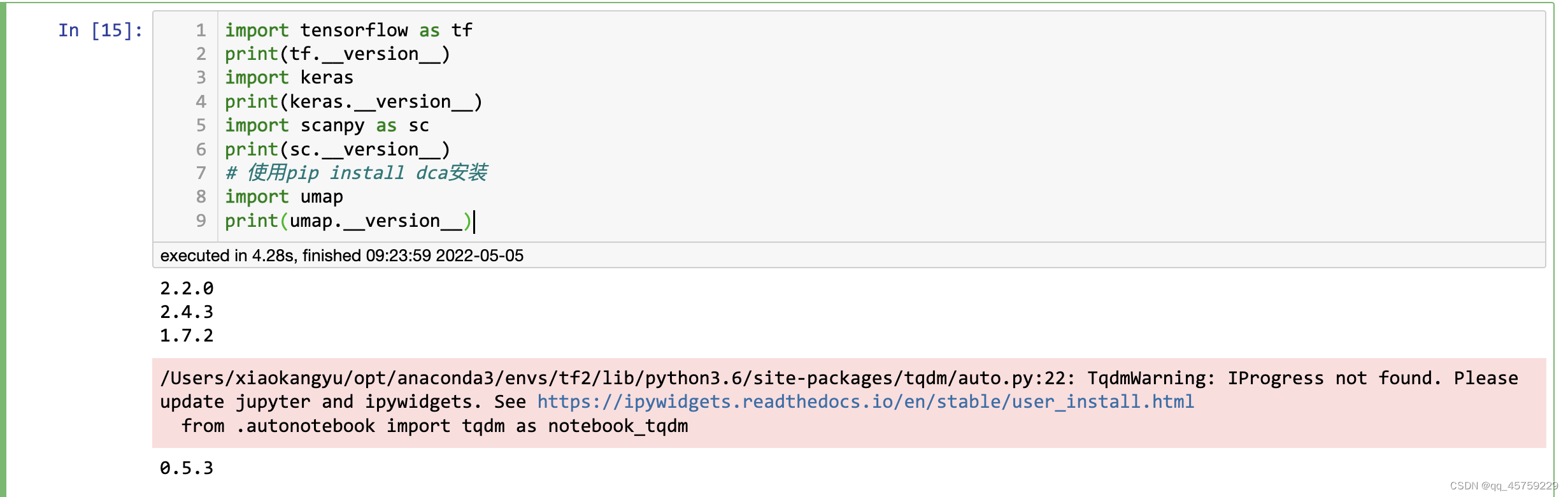
%load_ext rpy2.ipython
%%R -o counts -o meta
suppressMessages(library(splatter))
params <- newSplatParams()
params <- setParam(params, "nGenes", 5000)
params <- setParam(params, "batchCells", c(500,500,500))
params <- setParam(params, "batch.facLoc", 0.5)
params <- setParam(params, "batch.facScale", 0.5)
params <- setParam(params, "group.prob", c(1/3,1/3,1/3))
sim <- splatSimulate(params, method="groups", verbose=FALSE)
counts = data.frame(counts(sim))
meta = data.frame(colData(sim))
import anndata
import scanpy as sc
adata = anndata.AnnData(X=counts.values.T, obs=meta)
sc.pp.normalize_per_cell(adata,counts_per_cell_after=10000)
sc.pp.log1p(adata)
sc.tl.pca(adata,svd_solver='arpack')
sc.pp.neighbors(adata, n_neighbors=9)
sc.tl.umap(adata)
sc.pl.umap(adata,color=['Batch','Group'])
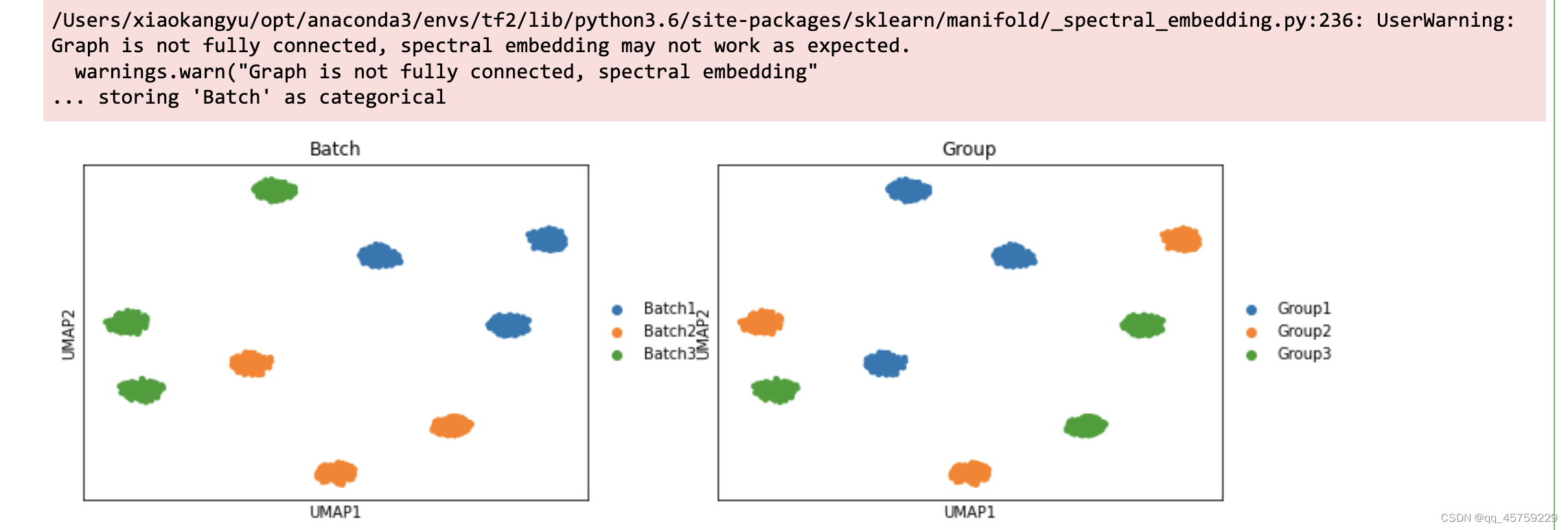
结果如下
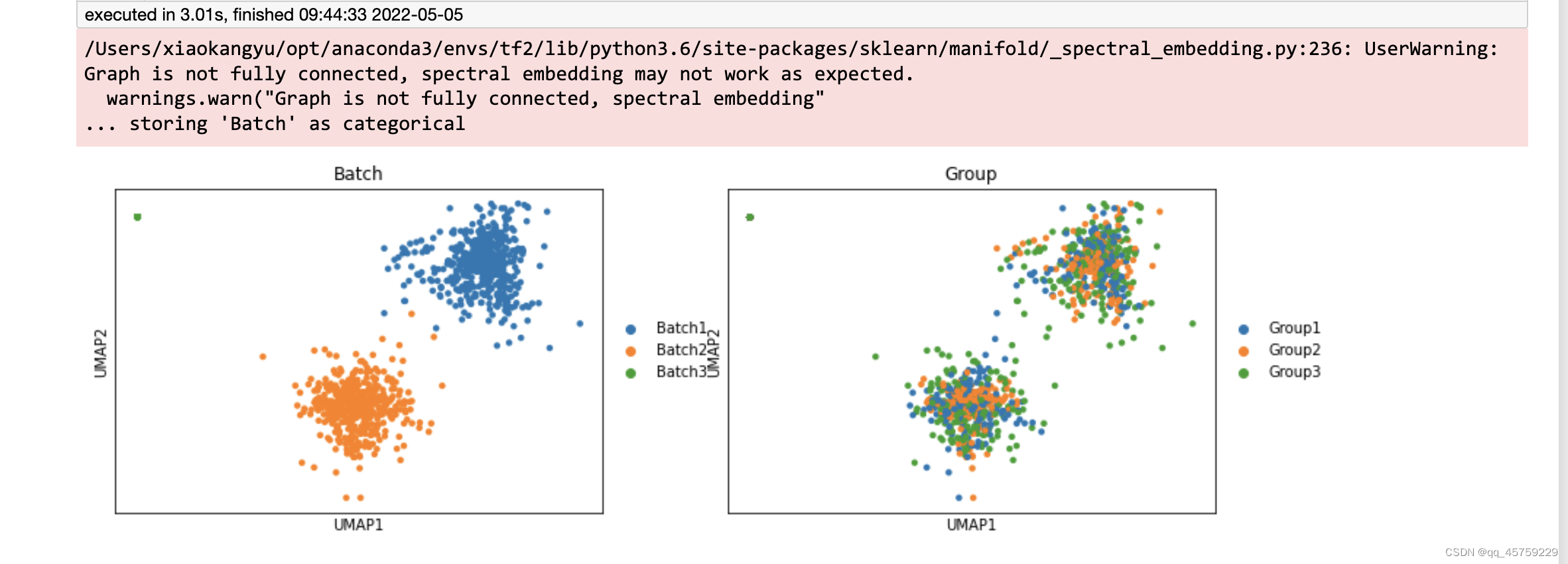 这个图很明显是不对的,而且louvain也没有安装
这个图很明显是不对的,而且louvain也没有安装
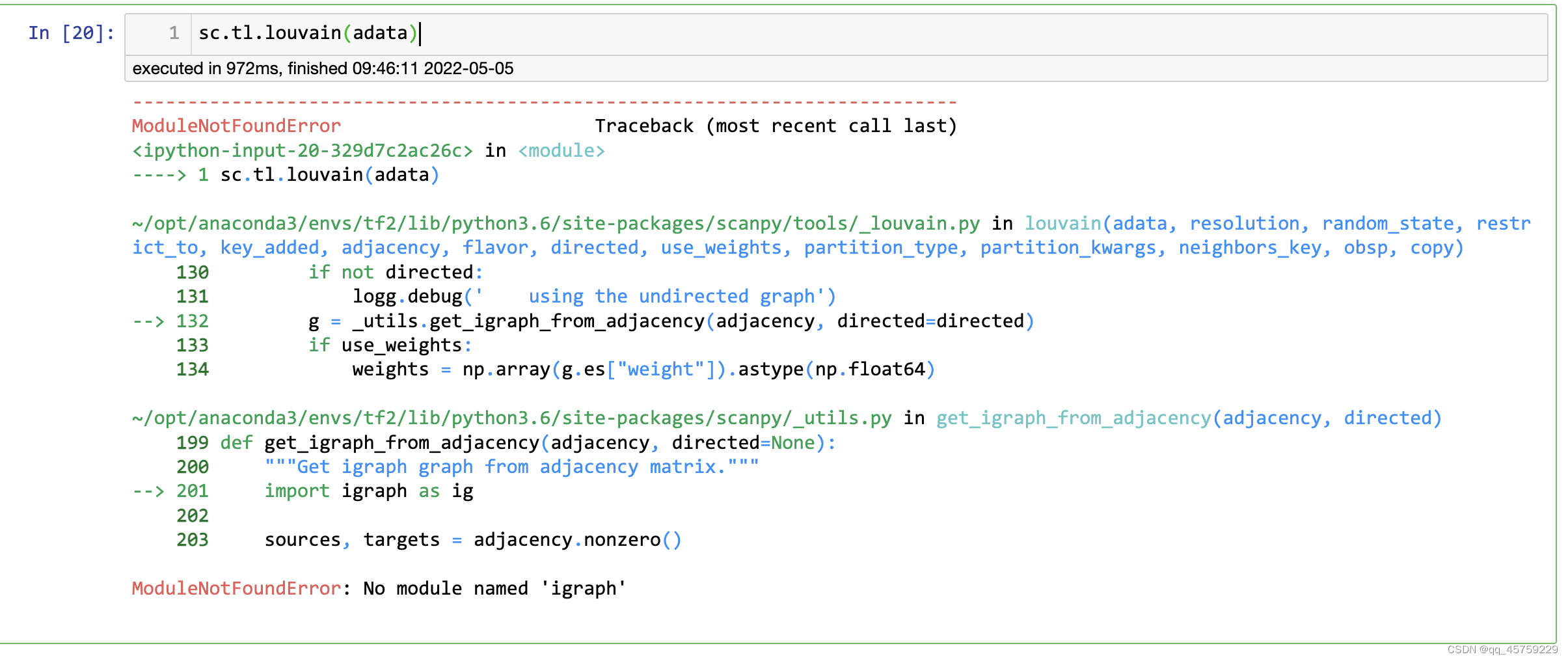
解决方法如下
conda install -c vtraag louvain # 可选,因为louvain 也要安装的
pip uninstall umap-learn
pip install umap-learn==0.4.6
pip uninstall numba
pip install numba==0.51.2
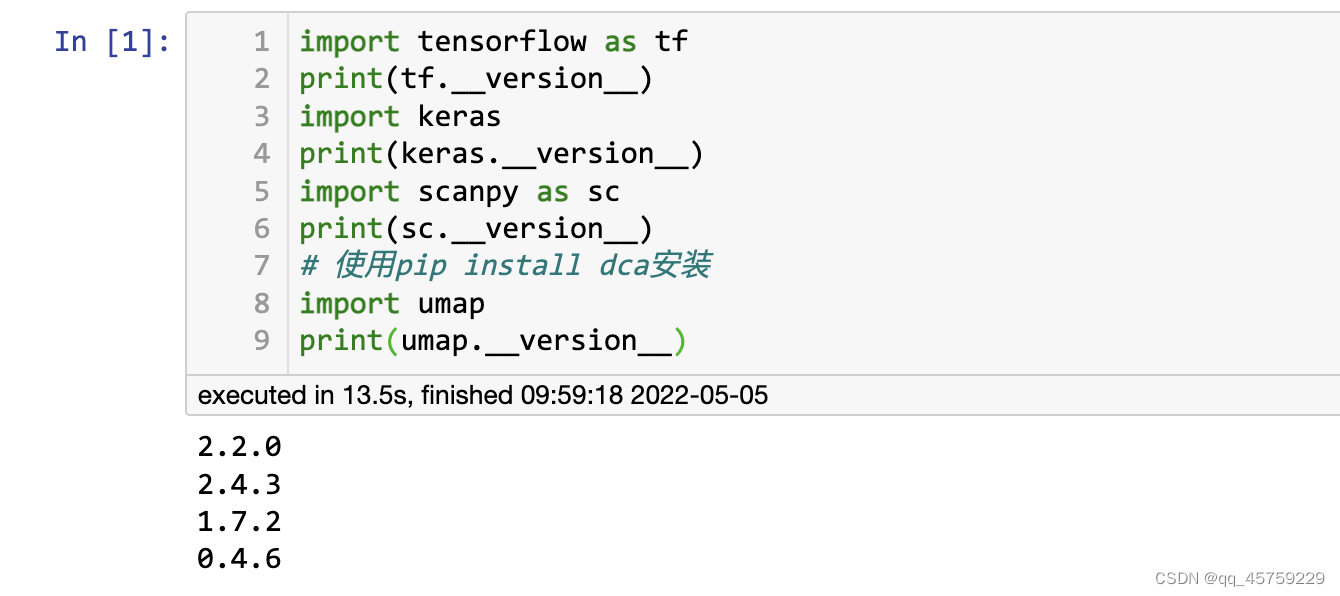 重新测试代码
重新测试代码
%load_ext rpy2.ipython
%%R -o counts -o meta
suppressMessages(library(splatter))
params <- newSplatParams()
params <- setParam(params, "nGenes", 5000)
params <- setParam(params, "batchCells", c(500,500,500))
params <- setParam(params, "batch.facLoc", 0.5)
params <- setParam(params, "batch.facScale", 0.5)
params <- setParam(params, "group.prob", c(1/3,1/3,1/3))
sim <- splatSimulate(params, method="groups", verbose=FALSE)
counts = data.frame(counts(sim))
meta = data.frame(colData(sim))
import anndata
import scanpy as sc
adata = anndata.AnnData(X=counts.values.T, obs=meta)
sc.pp.normalize_per_cell(adata,counts_per_cell_after=10000)
sc.pp.log1p(adata)
sc.tl.pca(adata,svd_solver='arpack')
sc.pp.neighbors(adata, n_neighbors=9)
sc.tl.umap(adata)
sc.pl.umap(adata,color=['Batch','Group'])
结果如下
 # 然后进行louvain
# 然后进行louvain
sc.tl.louvain(adata)
sc.pl.umap(adata,color=["louvain"])
结果如下
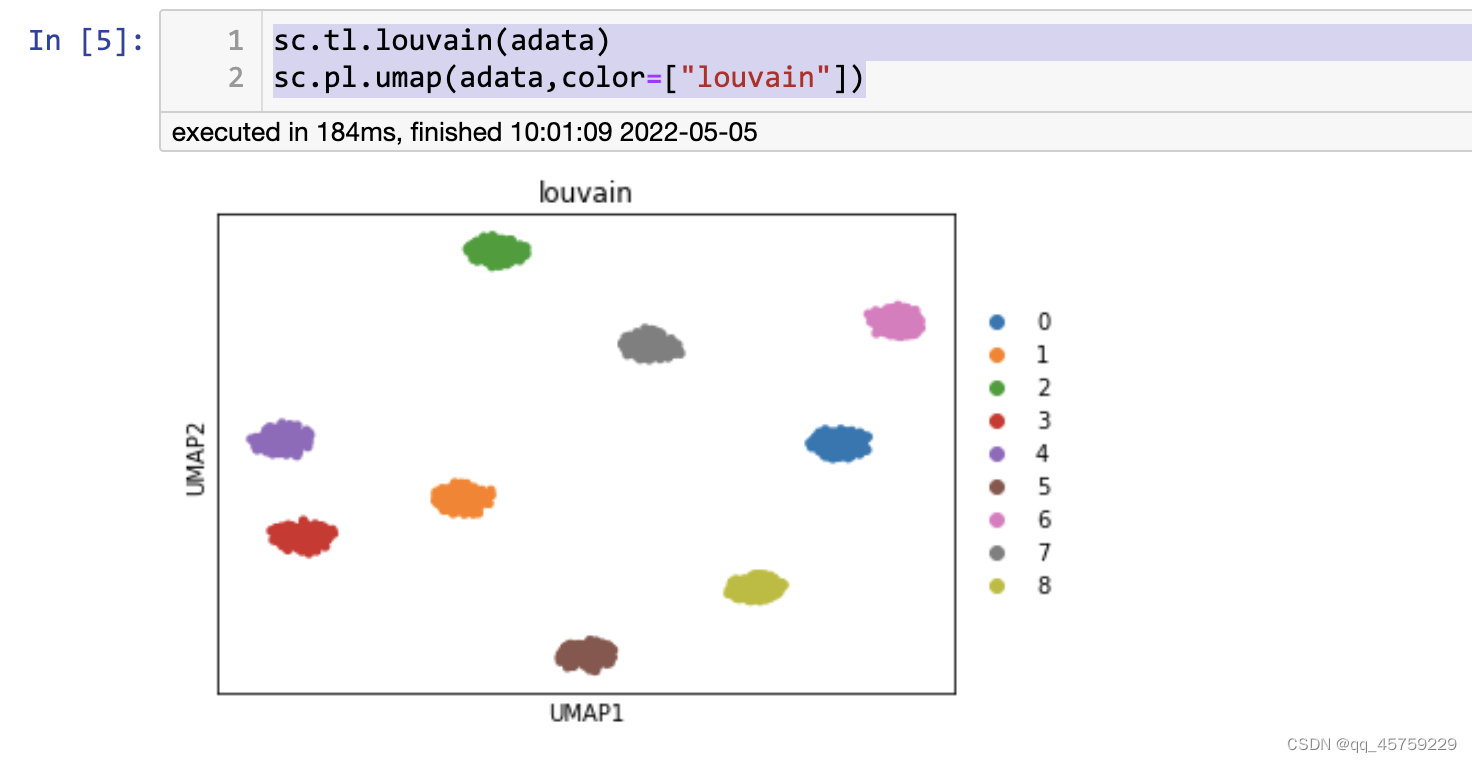 这个结果才是对的。
这个结果才是对的。
