import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn.cluster import KMeans
from sklearn.metrics import silhouette_score
from sklearn.preprocessing import MinMaxScaler
iris = pd.read_csv("E:/练习/Iris.csv")
x = iris.iloc[:, [0, 1, 2, 3]].values
iris.info()
iris[0:10]
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 150 entries, 0 to 149
Data columns (total 6 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 id 150 non-null int64
1 SepalLengthCm 150 non-null float64
2 SepalWidthCm 150 non-null float64
3 PetalLengthCm 150 non-null float64
4 PetalWidthCm 150 non-null float64
5 Species 150 non-null int64
dtypes: float64(4), int64(2)
memory usage: 7.2 KB
| id | SepalLengthCm | SepalWidthCm | PetalLengthCm | PetalWidthCm | Species |
|---|
| 0 | 1 | 5.1 | 3.5 | 1.4 | 0.2 | 0 |
|---|
| 1 | 2 | 4.9 | 3.0 | 1.4 | 0.2 | 0 |
|---|
| 2 | 3 | 4.7 | 3.2 | 1.3 | 0.2 | 0 |
|---|
| 3 | 4 | 4.6 | 3.1 | 1.5 | 0.2 | 0 |
|---|
| 4 | 5 | 5.0 | 3.6 | 1.4 | 0.2 | 0 |
|---|
| 5 | 6 | 5.4 | 3.9 | 1.7 | 0.4 | 0 |
|---|
| 6 | 7 | 4.6 | 3.4 | 1.4 | 0.3 | 0 |
|---|
| 7 | 8 | 5.0 | 3.4 | 1.5 | 0.2 | 0 |
|---|
| 8 | 9 | 4.4 | 2.9 | 1.4 | 0.2 | 0 |
|---|
| 9 | 10 | 4.9 | 3.1 | 1.5 | 0.1 | 0 |
|---|
iris.drop('id',axis=1,inplace=True)
iris.info()
iris[0:10]
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 150 entries, 0 to 149
Data columns (total 5 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 SepalLengthCm 150 non-null float64
1 SepalWidthCm 150 non-null float64
2 PetalLengthCm 150 non-null float64
3 PetalWidthCm 150 non-null float64
4 Species 150 non-null int64
dtypes: float64(4), int64(1)
memory usage: 6.0 KB
| SepalLengthCm | SepalWidthCm | PetalLengthCm | PetalWidthCm | Species |
|---|
| 0 | 5.1 | 3.5 | 1.4 | 0.2 | 0 |
|---|
| 1 | 4.9 | 3.0 | 1.4 | 0.2 | 0 |
|---|
| 2 | 4.7 | 3.2 | 1.3 | 0.2 | 0 |
|---|
| 3 | 4.6 | 3.1 | 1.5 | 0.2 | 0 |
|---|
| 4 | 5.0 | 3.6 | 1.4 | 0.2 | 0 |
|---|
| 5 | 5.4 | 3.9 | 1.7 | 0.4 | 0 |
|---|
| 6 | 4.6 | 3.4 | 1.4 | 0.3 | 0 |
|---|
| 7 | 5.0 | 3.4 | 1.5 | 0.2 | 0 |
|---|
| 8 | 4.4 | 2.9 | 1.4 | 0.2 | 0 |
|---|
| 9 | 4.9 | 3.1 | 1.5 | 0.1 | 0 |
|---|
iris_outcome = pd.crosstab(index=iris['Species'],
columns="count")
iris_outcome
| col_0 | count |
|---|
| Species | |
|---|
| 0 | 50 |
|---|
| 1 | 50 |
|---|
| 2 | 50 |
|---|
iris_setosa=iris.loc[iris["Species"]==0]
iris_virginica=iris.loc[iris["Species"]==1]
iris_versicolor=iris.loc[iris["Species"]==2]
sns.FacetGrid(iris,hue="Species",size=3).map(sns.distplot,"PetalLengthCm").add_legend()
sns.FacetGrid(iris,hue="Species",size=3).map(sns.distplot,"PetalWidthCm").add_legend()
sns.FacetGrid(iris,hue="Species",size=3).map(sns.distplot,"SepalLengthCm").add_legend()
plt.show()
D:\Anaconda3\lib\site-packages\seaborn\distributions.py:2557: FutureWarning: `distplot` is a deprecated function and will be removed in a future version. Please adapt your code to use either `displot` (a figure-level function with similar flexibility) or `histplot` (an axes-level function for histograms).
warnings.warn(msg, FutureWarning)
D:\Anaconda3\lib\site-packages\seaborn\distributions.py:2557: FutureWarning: `distplot` is a deprecated function and will be removed in a future version. Please adapt your code to use either `displot` (a figure-level function with similar flexibility) or `histplot` (an axes-level function for histograms).
warnings.warn(msg, FutureWarning)
D:\Anaconda3\lib\site-packages\seaborn\distributions.py:2557: FutureWarning: `distplot` is a deprecated function and will be removed in a future version. Please adapt your code to use either `displot` (a figure-level function with similar flexibility) or `histplot` (an axes-level function for histograms).
warnings.warn(msg, FutureWarning)
D:\Anaconda3\lib\site-packages\seaborn\axisgrid.py:316: UserWarning: The `size` parameter has been renamed to `height`; please update your code.
warnings.warn(msg, UserWarning)
D:\Anaconda3\lib\site-packages\seaborn\distributions.py:2557: FutureWarning: `distplot` is a deprecated function and will be removed in a future version. Please adapt your code to use either `displot` (a figure-level function with similar flexibility) or `histplot` (an axes-level function for histograms).
warnings.warn(msg, FutureWarning)
D:\Anaconda3\lib\site-packages\seaborn\distributions.py:2557: FutureWarning: `distplot` is a deprecated function and will be removed in a future version. Please adapt your code to use either `displot` (a figure-level function with similar flexibility) or `histplot` (an axes-level function for histograms).
warnings.warn(msg, FutureWarning)
D:\Anaconda3\lib\site-packages\seaborn\distributions.py:2557: FutureWarning: `distplot` is a deprecated function and will be removed in a future version. Please adapt your code to use either `displot` (a figure-level function with similar flexibility) or `histplot` (an axes-level function for histograms).
warnings.warn(msg, FutureWarning)
D:\Anaconda3\lib\site-packages\seaborn\axisgrid.py:316: UserWarning: The `size` parameter has been renamed to `height`; please update your code.
warnings.warn(msg, UserWarning)
D:\Anaconda3\lib\site-packages\seaborn\distributions.py:2557: FutureWarning: `distplot` is a deprecated function and will be removed in a future version. Please adapt your code to use either `displot` (a figure-level function with similar flexibility) or `histplot` (an axes-level function for histograms).
warnings.warn(msg, FutureWarning)
D:\Anaconda3\lib\site-packages\seaborn\distributions.py:2557: FutureWarning: `distplot` is a deprecated function and will be removed in a future version. Please adapt your code to use either `displot` (a figure-level function with similar flexibility) or `histplot` (an axes-level function for histograms).
warnings.warn(msg, FutureWarning)
D:\Anaconda3\lib\site-packages\seaborn\distributions.py:2557: FutureWarning: `distplot` is a deprecated function and will be removed in a future version. Please adapt your code to use either `displot` (a figure-level function with similar flexibility) or `histplot` (an axes-level function for histograms).
warnings.warn(msg, FutureWarning)



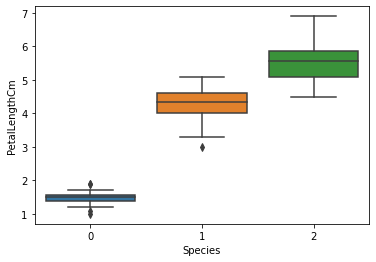
sns.boxplot(x="Species",y="PetalLengthCm",data=iris)
plt.show()
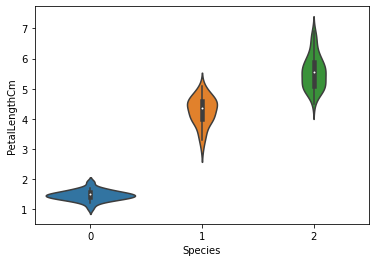
sns.violinplot(x="Species",y="PetalLengthCm",data=iris)
plt.show()
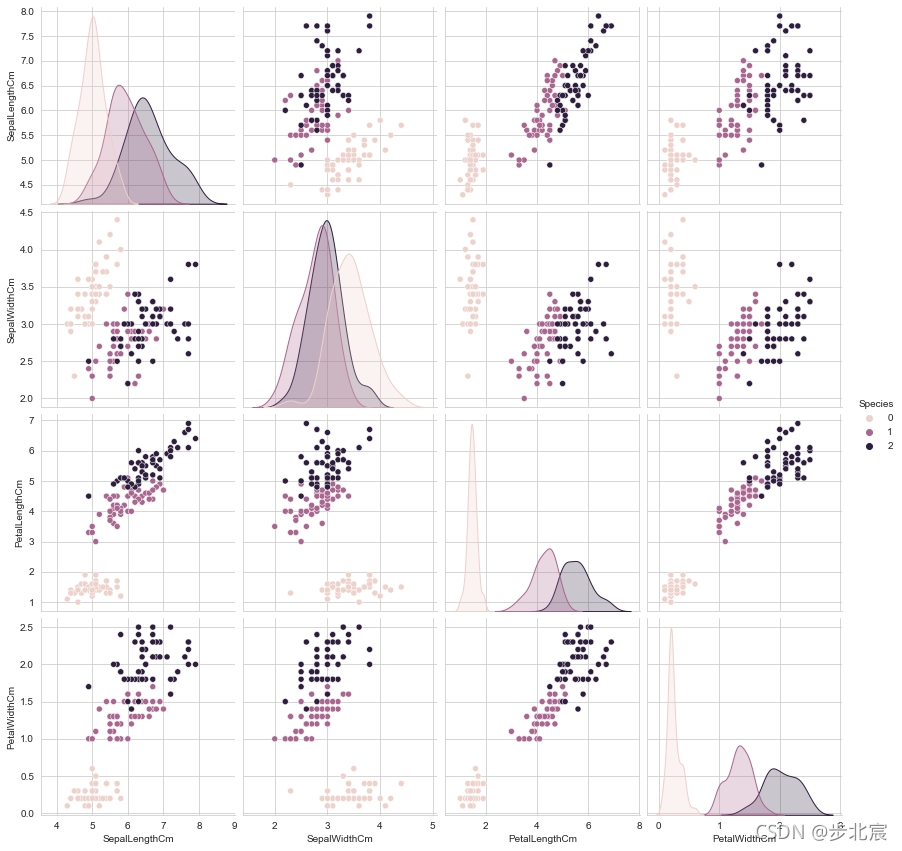
sns.set_style("whitegrid")
sns.pairplot(iris,hue="Species",size=3);
plt.show()
D:\Anaconda3\lib\site-packages\seaborn\axisgrid.py:1969: UserWarning: The `size` parameter has been renamed to `height`; please update your code.
warnings.warn(msg, UserWarning)
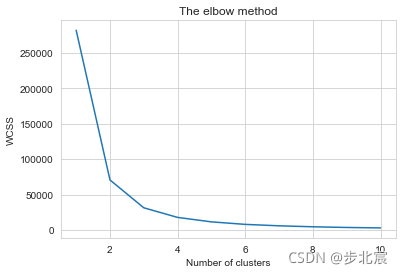
from sklearn.cluster import KMeans
wcss = []
for i in range(1, 11):
kmeans = KMeans(n_clusters = i, init = 'k-means++', max_iter = 300, n_init = 10, random_state = 0)
kmeans.fit(x)
wcss.append(kmeans.inertia_)
D:\Anaconda3\lib\site-packages\sklearn\cluster\_kmeans.py:881: UserWarning: KMeans is known to have a memory leak on Windows with MKL, when there are less chunks than available threads. You can avoid it by setting the environment variable OMP_NUM_THREADS=1.
warnings.warn(
plt.plot(range(1, 11), wcss)
plt.title('The elbow method')
plt.xlabel('Number of clusters')
plt.ylabel('WCSS')
plt.show()
kmeans = KMeans(n_clusters = 3, init = 'k-means++', max_iter = 300, n_init = 10, random_state = 0)
y_kmeans = kmeans.fit_predict(x)
plt.scatter(x[y_kmeans == 0, 0], x[y_kmeans == 0, 1], s = 100, c = 'purple', label = 'Iris-setosa')
plt.scatter(x[y_kmeans == 1, 0], x[y_kmeans == 1, 1], s = 100, c = 'orange', label = 'Iris-versicolour')
plt.scatter(x[y_kmeans == 2, 0], x[y_kmeans == 2, 1], s = 100, c = 'green', label = 'Iris-virginica')
plt.scatter(kmeans.cluster_centers_[:, 0], kmeans.cluster_centers_[:,1], s = 100, c = 'red', label = 'Centroids')
plt.legend()
<matplotlib.legend.Legend at 0x1f042341580>
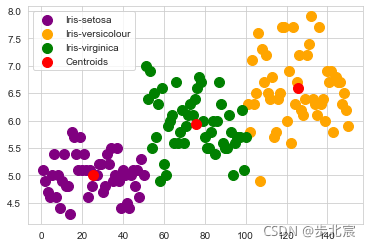
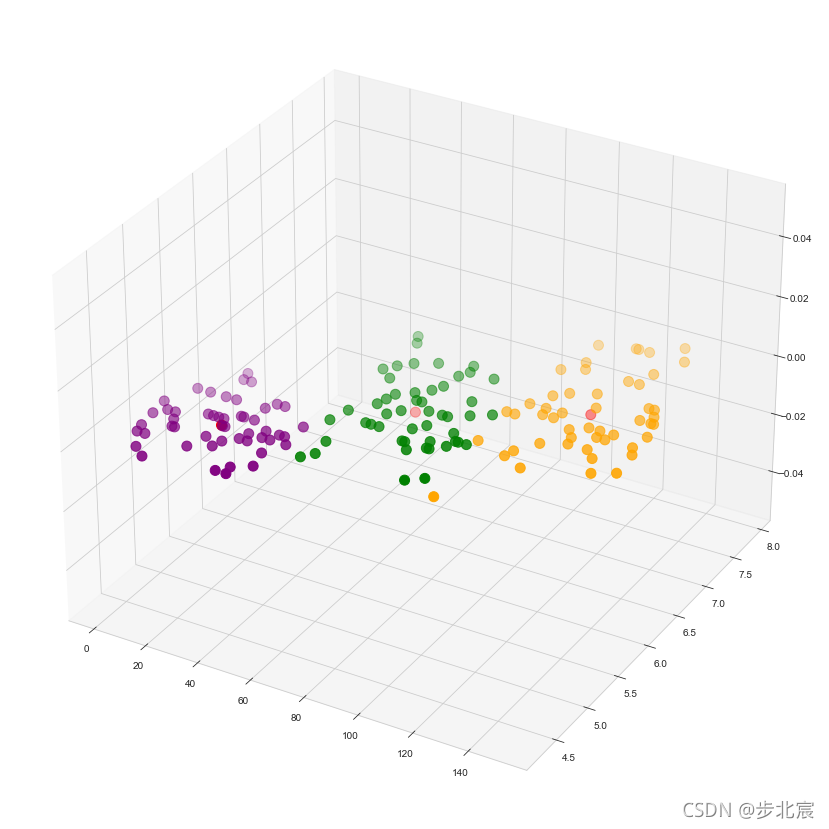
fig = plt.figure(figsize = (15,15))
ax = fig.add_subplot(111, projection='3d')
plt.scatter(x[y_kmeans == 0, 0], x[y_kmeans == 0, 1], s = 100, c = 'purple', label = 'Iris-setosa')
plt.scatter(x[y_kmeans == 1, 0], x[y_kmeans == 1, 1], s = 100, c = 'orange', label = 'Iris-versicolour')
plt.scatter(x[y_kmeans == 2, 0], x[y_kmeans == 2, 1], s = 100, c = 'green', label = 'Iris-virginica')
plt.scatter(kmeans.cluster_centers_[:, 0], kmeans.cluster_centers_[:,1], s = 100, c = 'red', label = 'Centroids')
plt.show()
|








