1. DNA sequences alignment
两个DNA序列如下:
sequence_A = "AGGCTATCACCTGACCTCCAGGCCGATGCCC"
sequence_B = "TAGCTATCACGACCGCGGTCGATTTGCCCGAC"
2. 对齐代码
import numpy as np
import matplotlib.pyplot as plt
from matplotlib import rcParams
from collections import namedtuple
rcParams['font.family'] = 'serif'
rcParams['font.size'] = 5
match = 8
mismatch = -5
gap = -3
score_tuple = namedtuple('score_tuple', ('score', 'point_position'))
def init_array(x, y):
array = [[0] * y for _ in range(x)]
array[0][0] = score_tuple(0, None)
for j in range(1, y):
array[0][j] = score_tuple(gap * j, [0, j - 1])
for i in range(1, x):
array[i][0] = score_tuple(gap * i, [i - 1, 0])
return array
def compute(array, seq1, seq2):
row, col = len(seq2), len(seq1)
for i in range(1, row + 1):
for j in range(1, col + 1):
if seq1[j - 1] == seq2[i - 1]:
s = match
else:
s = mismatch
lu = [array[i - 1][j - 1].score+s, [i - 1, j - 1]]
left = [array[i - 1][j].score + gap, [i - 1, j]]
up = [array[i][j - 1].score + gap, [i, j - 1]]
max_choice = max([lu, left, up], key=lambda x: x[0])
score = max_choice[0]
point_position = max_choice[1]
array[i][j] = score_tuple(score, point_position)
return array
def backtrack(array, seq1, seq2):
s1 = []
s2 = []
row, col = len(seq2), len(seq1)
while array[row][col].score != 0:
i, j = array[row][col].point_position
if i + 1 == row and j + 1 == col:
s1.append(seq1[col - 1])
s2.append(seq2[row - 1])
row, col = i, j
elif row == i + 1 and col == j:
s1.append("-")
s2.append(seq2[i])
row, col = i, j
elif row == i and col == j + 1:
s1.append(seq1[j])
s2.append("-")
row, col = i, j
s1 = ''.join(s1[::-1])
s2 = ''.join(s2[::-1])
return s1, s2
def main(seq1, seq2):
x, y = len(seq2) + 1, len(seq1) + 1
array = compute(init_array(x, y), seq1, seq2)
value, total_value = [], []
for j in range(len(array)):
for i in range(len(array[j])):
value.append(array[j][i].score)
total_value.extend([value])
value = []
m = len(array)
n = len(array[0])
for column, row in enumerate(total_value):
plt.text(5, 10 * column + 15, seq2[column - 1], horizontalalignment='center', verticalalignment='center')
for line, num in enumerate(row):
if column == 0:
plt.text(10 * line + 15, 5, seq1[line - 1], horizontalalignment='center', verticalalignment='center')
plt.text(10 * line + 15, 10 * column + 15, num, horizontalalignment='center', verticalalignment='center')
plt.axis([0, 10 * (n + 1), 10 * (m + 1), 0])
plt.xticks(np.linspace(0, 10 * (n + 1), n + 2), [])
plt.yticks(np.linspace(0, 10 * (m + 1), m + 2), [])
plt.grid(linestyle="solid")
plt.show()
s1, s2 = backtrack(array, seq1, seq2)
max_score = array[x-1][y-1].score
print("最大得分:%d\n sequence1:%s\n sequence2:%s" % (max_score, s1, s2))
if __name__ == '__main__':
sequence_A = "AGGCTATCACCTGACCTCCAGGCCGATGCCC"
sequence_B = "TAGCTATCACGACCGCGGTCGATTTGCCCGAC"
main(sequence_A, sequence_B)
3. 图以及输出结果
绘图结果:

输出结果:
最大得分:149
sequence1:-AGGCTATCACCTGACCTCCAGGCCGA--TG-CC--C
sequence2:TA-GCTATCA-C-GACC-GC-GGTCGATTTGCCCGAC
手描图:
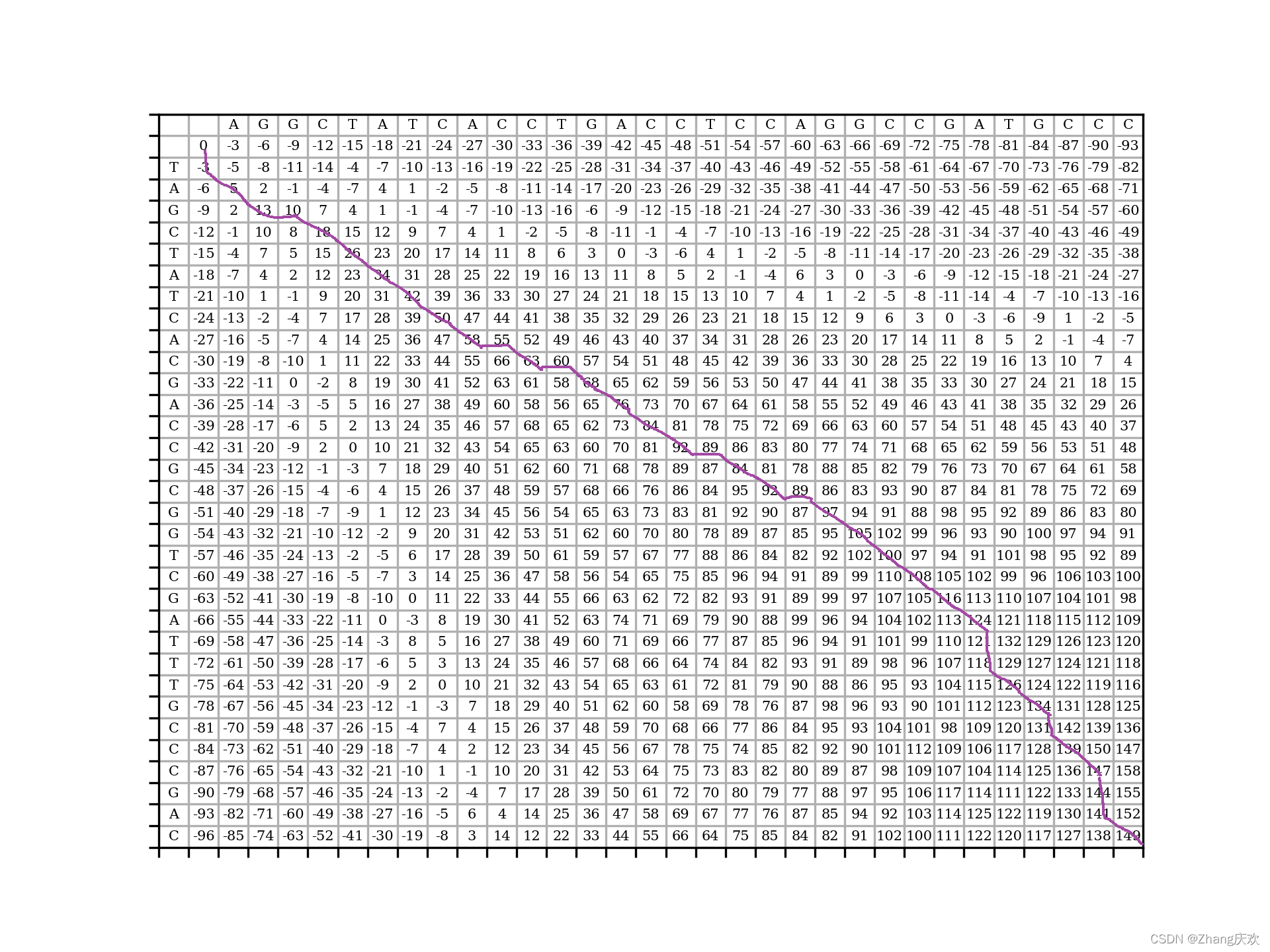
后面这个折线画起来太麻烦了,所以直接用手画了