机器学习入门-肝病预测分析
导入函数和支持包
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import StandardScaler
from sklearn.ensemble import IsolationForest
1、数据描述
该数据集包含416条肝脏患者记录和167条非肝脏患者记录,从印度安得拉邦东北部收集的。选择器是用于划分为组(肝脏患者与否)的类标签。该数据集包含441份男性患者记录和142份女性患者记录。任何年龄超过89岁的患者都被列为年龄"90"。
下载地址:UCI数据库
2、载入数据
因为数据集的特征与数据部分是分开的,因此我们需要人为增加上去。
data_col=['age', 'gender', 'total Bilirubin', 'direct Bilirubin'
, 'total proteins', 'albumin', 'A/G ratio', 'SGPT','SGOT','Alkphos','Type']
data=pd.read_csv('Indian LiverPatient.csv',names=data_col)#加入你自己的路径,或者放在同一文件夹下也可
3、查看Type类型的数量
data['Type'].value_counts()#416条肝脏患者记录和167条非肝脏患者记录

4、检查是否存在空值
data.isnull().sum()
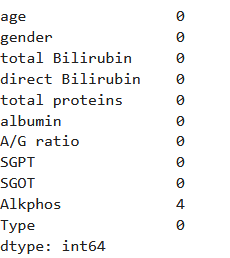
可以看到Alkphos中存在空值
5、查看描述性统计量
data.describe()
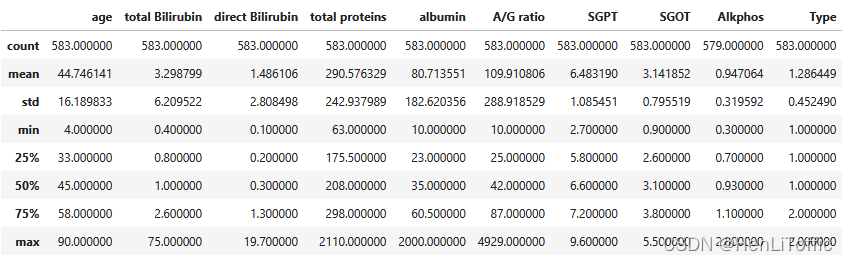
对于上面的空值简单采取平均数填充
data=data.fillna(0.930000)
删除data中的重复值
data=data.drop_duplicates()
对于gender进行哑变量处理,机器学习中不支持中文或者空值的出现
dummy_gender= pd.get_dummies(data.iloc[:,1],prefix = "gender")
data= pd.concat([data,dummy_gender.iloc[:,0:2]], axis= 1)
data_new=data.drop(['gender'],axis=1)#获取新的特征并合并到原来的表上,并删除原来的gender列
6、画出热力图并分析特征相关性
def corr_map(df):
var_corr = df.corr()
mask = np.zeros_like(var_corr, dtype=np.bool)
mask[np.triu_indices_from(mask)] = True
cmap = sns.diverging_palette(220, 10, as_cmap=True)
f, ax = plt.subplots(figsize=(20, 12))
sns.set(font_scale=1)
sns.heatmap(var_corr, mask=mask, cmap=cmap, vmax=1, center=0
,square=True, linewidths=.5, cbar_kws={"shrink": .5}
,annot=True,annot_kws={'size':12,'weight':'bold', 'color':'c'})
plt.show()
corr_map(data_new)
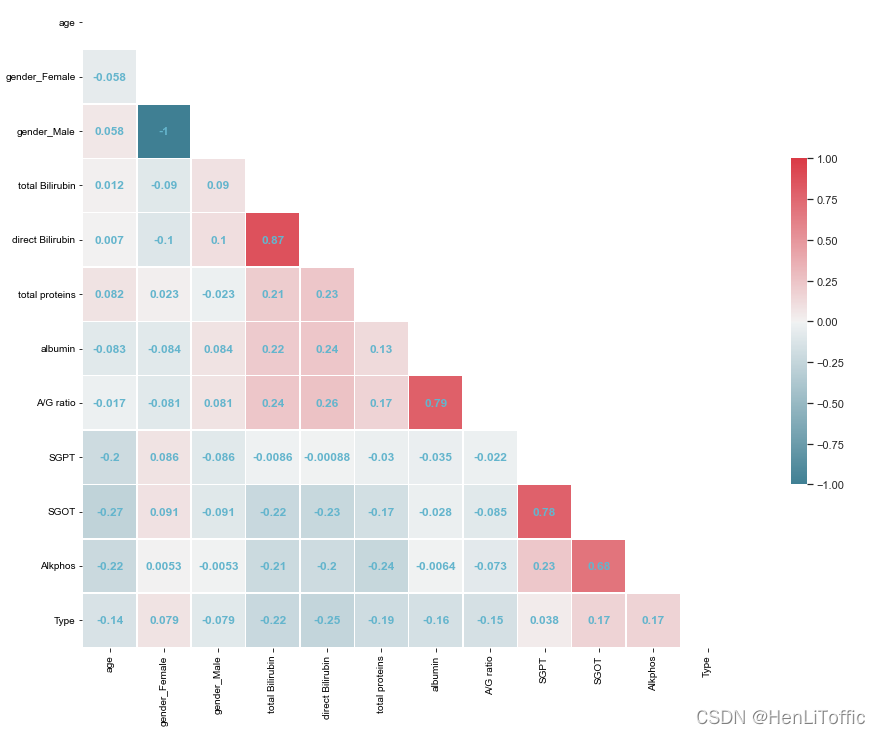
7、对数据进行标准化
from sklearn import preprocessing
from sklearn.preprocessing import StandardScaler
features = ['age','gender_Female', 'gender_Male','total Bilirubin', 'direct Bilirubin', 'total proteins',
'albumin', 'A/G ratio', 'SGPT', 'SGOT', 'Alkphos']
data_gyh= data_new[features]
data_gyh[features] = StandardScaler().fit_transform(data_gyh)
print(data_gyh.shape)
data_gyh.head()
8、划分数据集并进行模型的训练
X, y = data_new.iloc[:, 1:-1], data_new.iloc[:, -1] # 把数据与标签拆分开来
X_train, X_test, y_train, y_test = train_test_split(X, y,test_size=0.3, random_state=0)
from sklearn.ensemble import RandomForestClassifier# 使用随机森林分类
clf = RandomForestClassifier()
clf.fit(X_train,y_train)
print(clf)
from sklearn import metrics
y_predict = clf.predict(X_test)
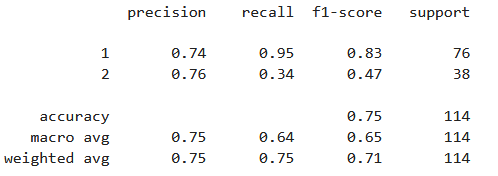
print(metrics.classification_report(y_test, y_predict))#输出模型的报告
9、模型调参
from sklearn.model_selection import GridSearchCV
from sklearn.model_selection import cross_val_score
调整树的棵树参数得到最佳参数
scorel = []
for i in range(0,200,10):
rfc = RandomForestClassifier(n_estimators=i+1,
n_jobs=-1,
random_state=90)
score = cross_val_score(rfc,X_train,y_train,cv=10).mean()
scorel.append(score)
print(max(scorel),(scorel.index(max(scorel))*10)+1)
plt.figure(figsize=[20,5])
plt.plot(range(1,201,10),scorel)
plt.show()

调整分割内部节点所需的最小样本数
scorel = []
for i in range(3,50):
rfc = RandomForestClassifier(n_estimators=71,
min_samples_split=i,
n_jobs=-1,
random_state=90)
score = cross_val_score(rfc,X_train,y_train,cv=10).mean()
scorel.append(score)
print(max(scorel),([*range(3,50)][scorel.index(max(scorel))]))
plt.figure(figsize=[20,5])
plt.plot(range(3,50),scorel)
plt.show()

10、输出特征重要性
importances = clf.feature_importances_
#计算随机森林中所有的树的每个变量的重要性的标准差
std = np.std([tree.feature_importances_ for tree in clf.estimators_],axis=0)
#按照变量的重要性排序后的索引
indices = np.argsort(importances)[::-1]
plt.figure()
plt.figure(figsize=(10,5))
plt.title("Feature importances")
plt.bar(range(X_train.shape[1]), importances[indices], color="c", yerr=std[indices], align="center")
plt.xticks(fontsize=14)
plt.xticks(range(X_train.shape[1]),data_new.columns.values[:-1][indices],rotation=40)
plt.xlim([-1,X_train.shape[1]])
plt.tight_layout()
plt.show()
