参考:https://github.com/PaddlePaddle/PaddleHelix/blob/dev/tutorials/README_cn.md
前提先安装pahelix的一个独立conda环境
1、化合物分子
下载预训练模型,修改model网络输出
C:\Users\lonng\paddlehelix\PaddleHelix\apps\pretrained_compound\pretrain_gnns\src\model.py
import os
import numpy as np
import sys
sys.path.insert(0, os.getcwd() + "/..")
os.chdir(r"C:\Use***helix\PaddleHelix\apps\pretrained_compound\pretrain_gnns")
import paddle
import paddle.nn as nn
import paddle.distributed as dist
import pgl
from pahelix.model_zoo.pretrain_gnns_model import PretrainGNNModel, AttrmaskModel
from pahelix.datasets.zinc_dataset import load_zinc_dataset
from pahelix.utils.splitters import RandomSplitter
from pahelix.featurizers.pretrain_gnn_featurizer import AttrmaskTransformFn, AttrmaskCollateFn
from pahelix.utils import load_json_config
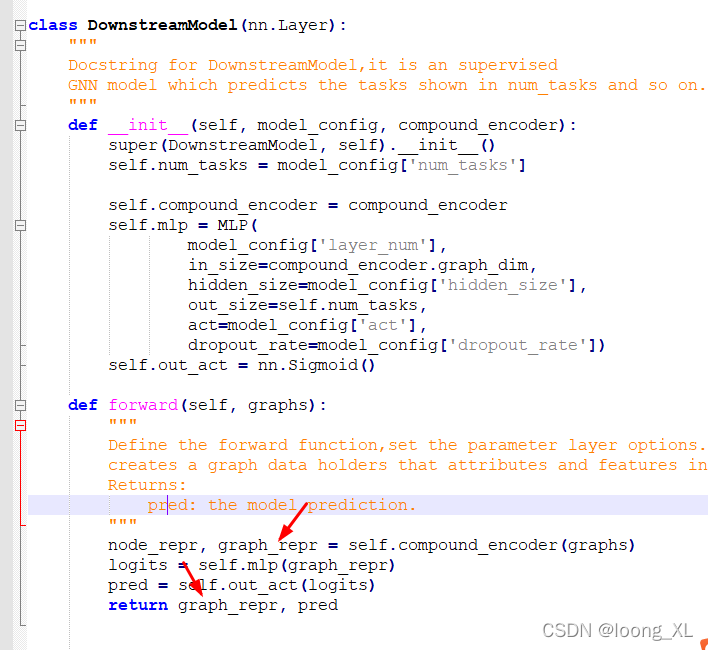
from src.model import DownstreamModel
from src.featurizer import DownstreamTransformFn, DownstreamCollateFn
from src.utils import calc_rocauc_score, exempt_parameters
task_names = ['NR-AR', 'NR-AR-LBD', 'NR-AhR', 'NR-Aromatase', 'NR-ER', 'NR-ER-LBD', 'NR-PPAR-gamma', 'SR-ARE', 'SR-ATAD5', 'SR-HSE', 'SR-MMP', 'SR-p53']
compound_encoder_config = load_json_config(r"C:\Use***ehelix\PaddleHelix\apps\pretrained_compound\pretrain_gnns\model_configs\pregnn_paper.json")
model_config = load_json_config(r"C:\User***dlehelix\PaddleHelix\apps\pretrained_compound\pretrain_gnns\model_configs\down_linear.json")
model_config['num_tasks'] = len(task_names)
compound_encoder = PretrainGNNModel(compound_encoder_config)
model = DownstreamModel(model_config, compound_encoder)
model.set_state_dict(paddle.load(r'C:\Users\lonng\paddlehelix\pregnn-attrmask-supervised'))
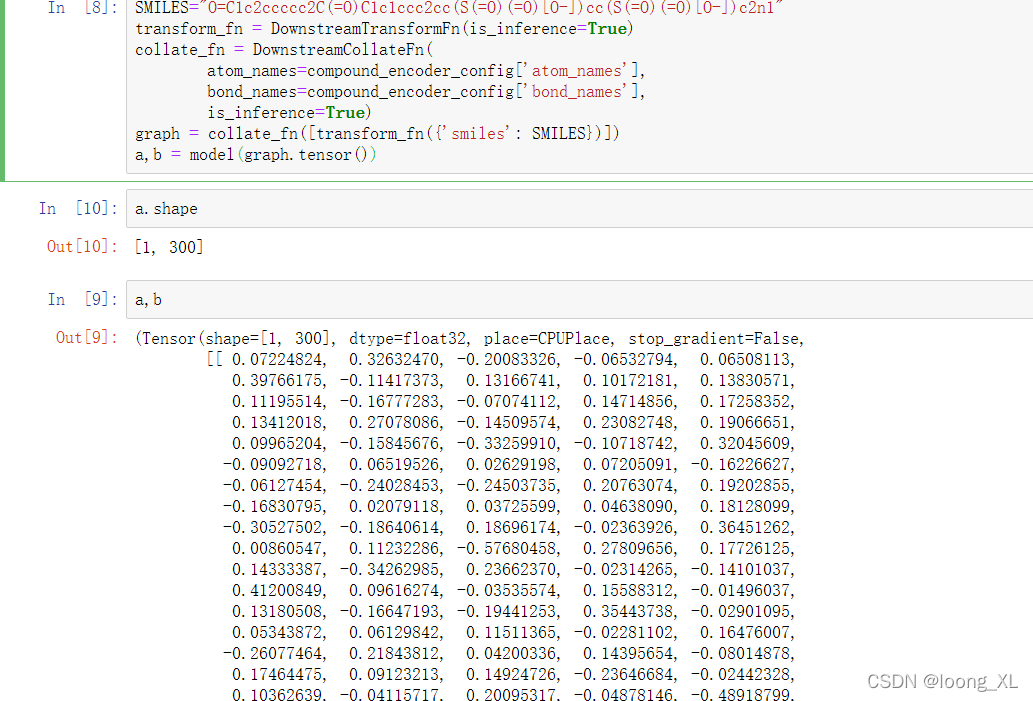
SMILES="O=C1c2ccccc2C(=O)C1c1ccc2cc(S(=O)(=O)[O-])cc(S(=O)(=O)[O-])c2n1"
transform_fn = DownstreamTransformFn(is_inference=True)
collate_fn = DownstreamCollateFn(
atom_names=compound_encoder_config['atom_names'],
bond_names=compound_encoder_config['bond_names'],
is_inference=True)
graph = collate_fn([transform_fn({'smiles': SMILES})])
a,b = model(graph.tensor())
2、蛋白质向量表示
import os
import time
import sys
import argparse
import json
import codecs
import numpy as np
import random
import paddle
import paddle.nn.functional as F
from pahelix.model_zoo.protein_sequence_model import ProteinEncoderModel, ProteinModel, ProteinCriterion
from pahelix.utils.protein_tools import ProteinTokenizer
predict_model = r'C:\Users\lonng\paddlehelix\tape_resnet_pretrain.pdparam'
paddle.set_device("cpu")
model_config = \
{
"model_name": "secondary_structure",
"task": "seq_classification",
"class_num": 3,
"label_name": "labels3",
"model_type": "resnet",
"hidden_size": 512,
"layer_num": 3,
"comment": "The following hyper-parameters are optional.",
"dropout_rate": 0.1,
"weight_decay": 0.01
}
import os
import numpy as np
import sys
sys.path.insert(0, os.getcwd() + "/..")
os.chdir(r"C:\Users\lonng\paddlehelix\PaddleHelix\apps\pretrained_protein\tape")
from data_gen import create_dataloader, pad_to_max_seq_len
from metrics import get_metric
from paddle.distributed import fleet
encoder_model = ProteinEncoderModel(model_config, name='protein')
model = ProteinModel(encoder_model, model_config)
model.load_dict(paddle.load(predict_model))
tokenizer = ProteinTokenizer()
examples = [
'MVLSPADKTNVKAAWGKVGAHAGEYGAEALERMFLSFPTTKTYFPHFDLSHGSAQVKGHGKKVADALTNAVAHVDDMPNALSALSDLHAHKLRVDPVNFKLLSHCLLVTLAAHLPAEFTPAVHASLDKFLASVSTVLTSKYR',
'KQHTSRGYLHEFDGDPANRCHQSLYKWHDKDCDWLVDWEMKPMDALMETDHQPSMLVHLEQSYKWFCCIKGKPLNFAALLDGWTKITPMAKALYWRDHISEAWLIQCMFEEKILIVRTLMDENGTHKNYFVMSRLCGSCITFEWDSWEAEKPHKVWMGMKNCVSWKRKDVIEMVFERTQWAKWADNIYNWACCPMQVPEIIPFQFFYQTDENFCFKLLMKPCKFYYFSCHHLGHLHCLLKYQWYKGVYLGMRLRVFHKMIVCFHGHWTWVEGNSGIEGRGGIMMHTGITMDCFFDRNIQQSYGGSRWSEQNMKHSQHSRCDPYRTCEPEGTTPEQKCVQRQRIKVRVCHMPEDCLWTSCV',
]
example_ids = [tokenizer.gen_token_ids(example) for example in examples]
max_seq_len = max([len(example_id) for example_id in example_ids])
pos = [list(range(1, len(example_id) + 1)) for example_id in example_ids]
pad_to_max_seq_len(example_ids, max_seq_len)
pad_to_max_seq_len(pos, max_seq_len)
texts = paddle.to_tensor(example_ids)
pos = paddle.to_tensor(pos)
encoder_repr = encoder_model(texts, pos)
print(encoder_repr.shape)
encoder_repr