基于Pytorch的卷积神经网络CNN实例应用及详解
一、卷积神经网络CNN定义
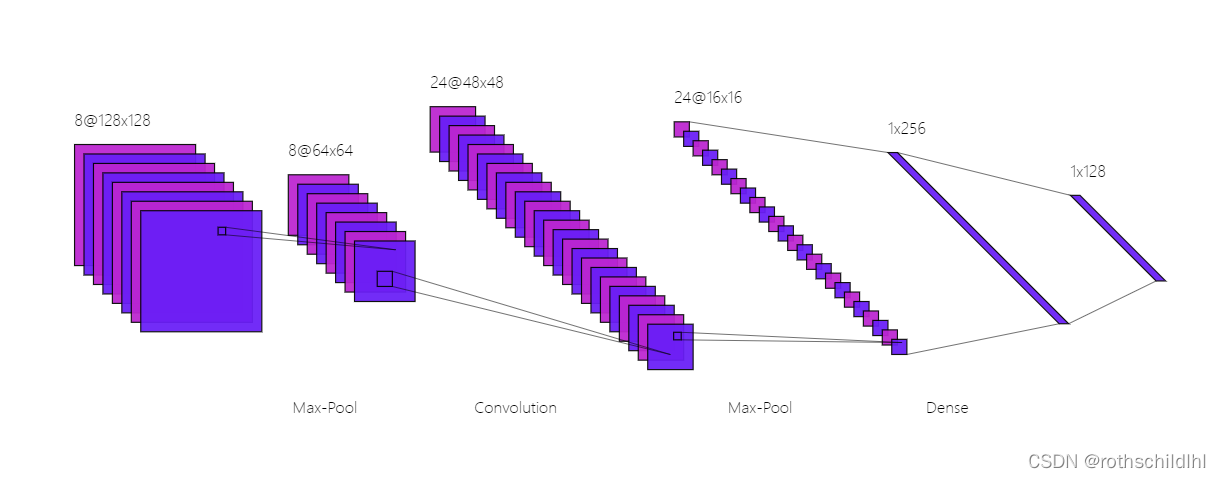
卷积神经网络(CNN,有时被称为 ConvNet)是很吸引人的。在短时间内,它们变成了一种颠覆性的技术,打破了从文本、视频到语音等多个领域所有最先进的算法,远远超出了其最初在图像处理的应用范围。CNN 由许多神经网络层组成。卷积和池化这两种不同类型的层通常是交替的。网络中每个滤波器的深度从左到右增加。最后通常由一个或多个全连接的层组成。
二、卷积神经网络CNN的原理
- 博主学习卷积神经网络CNN主要参考下面的两篇文章:
- 第一篇参考文章:点击打开《卷积神经网络推导和实现》文章
- 第二篇参考文章:点击打开《卷积神经网络(CNN)基础及经典模型介绍》文章
- 阅读上面两篇文章主要需要掌握CNN的数学原理公式,卷积、填充、池化、全连接等的概念。
三、卷积神经网络CNN实现的前期准备
- 下载编辑器和配置程序运行环境:点击打开《基于Windows中学习Deep Learning之搭建Anaconda+Pytorch(Cuda+Cudnn)+Pycharm工具和配置环境完整最简版》文章
- 项目实现用到的知识点如下:
- 点击打开《Pytorch实践中的list、numpy、torch.tensor之间数据格式的相互转换方法》文章
- 点击打开《Pytorch之nn.Conv1d学习个人见解》文章
- 点击打开《Pytorch中使用torch.nn模块进行神经网络模型初步构造》文章
- 点击打开《Pytorch中设计随机数种子的必要性》文章
- 点击打开《Pytorch中Trying to backward through the graph和one of the variables needed for gradient错误解决方案》文章
四、卷积神经网络CNN实现案例分析
-
案例目的:是构造卷积神经网络模型训练后进行药物靶体交互的二分类(0和1)预测。
-
数据集及格式说明:首先案例用到的所有数据集是来自KIBA 数据集,数据以txt文本数据存储。
-
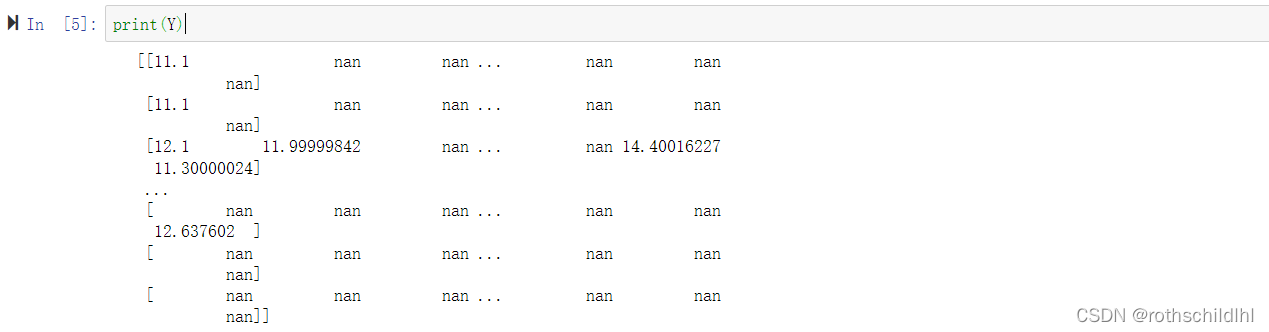
亲和矩阵数据集Y说明:亲和矩阵的维度是[229,2110],分别表示229种蛋白质和2110种药物分子,矩阵对应的数值表示对应蛋白质和药物分子交互的亲和性,若为“nan”表示无亲和性也就是后续分类为0,其余为1。
-
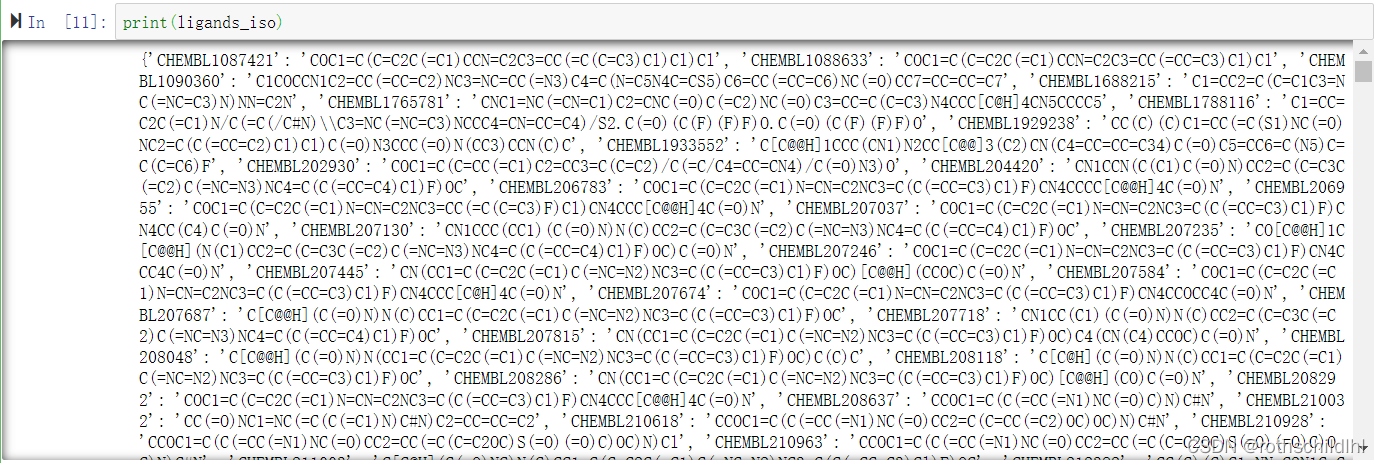
药物及其smiles序列数据集ligands_iso说明:药物名称序号和对应的结构smiles序列,以字典形式保存,长度是2111(表示有2111种药物)。
-
蛋白质及其sequences序列数据集proteins说明:蛋白质名称序号和对应的结构sequences序列,以字典形式保存,长度为229(表示有229种蛋白质)。

-
训练集train_fold_setting说明:数据维度是[5,19709],也就是分为五组,每组包含19709个数字,其中数字N表示为亲和矩阵数据集Y所有行列组合数目是483190(229*2110)种组合中按顺序排的第N组。
-
测试集test_fold_setting说明:数据维度是[19709],也就是只有一组,该组包含19709个数字,其中数字N表示为亲和矩阵数据集Y所有行列组合数目是483190(229*2110)种组合中按顺序排的第N组。
-
DTI案例CNN模型搭建大致如下(具体实现细节可见后面的代码):
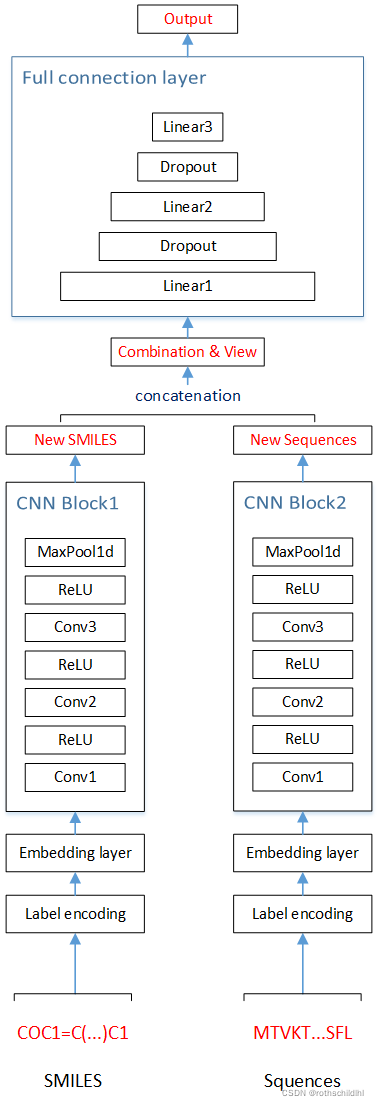
-
Label encoding 说明:将药物分子和蛋白质的序列的各种字符用数字字符替换,便于后面的词嵌入操作。
-
Embedding layer 说明:将药物分子和蛋白质的数字序列进行词嵌入操作,也就是增加一维度表示,比如原来的维度是一维只有长度(药物分子初始设置为100,蛋白质初始设置为1000),增加宽度(宽度初始设置都为128)这一维度。注意:卷积后长度宽度初始设置会变化。
-
concatenation(串联)说明:将原本分开的每组药物分子特征和蛋白质序列特征拼接起来,也就是根据宽度相同将两者的长拼接起来,所以拼接的宽度不变,长度两者相加。
五、卷积神经网络CNN实现完整代码和结果
import torch as t
import pickle
import numpy as np
import json
import math
from torch import nn
import torch.nn.functional as F
from sklearn.metrics import accuracy_score
# 构建训练集和测试集数据转换函数,对蛋白质和药物分子数据进行数据转换
def datatransform(data):
drug = [] # 训练集和测试集对应的各下标的药物(配体)
protein = [] # 训练集和测试集对应的各下标的蛋白质(靶体)
for i in range(len(data)):
drug.append(math.floor(data[i] / 229))
protein.append((data[i] % 229))
# 读取亲和矩阵Y数据
# 亲和矩阵Y的维度是[229,2110],229表示蛋白质的种类数量,2110表示药物分子的种类数量,矩阵元素的值表示蛋白质与药物分子结合的亲和性,nan值表示不亲和也就是互相无作用
Y = pickle.load(open("E:/data/kiba/Y", "rb"), encoding='latin1')
effective = [] # 训练集和测试集对应的各下标的药物和蛋白质结合的有效性(亲和性)
for i in range(len(data)):
d = drug[i]
p = protein[i]
if np.isnan(Y[d][p]): # 判断受体靶体有无亲和性
effective.append(0) # 受体靶体无亲和性
else:
effective.append(1) # 受体靶体有亲和性
# list转为tensor
effectives = t.LongTensor(effective) # 为损失函数计算做准备
# 读取药物和其smiles序列
drugs = json.load(open("E:\data\kiba\ligands_iso.txt"))
drug_smiles = [] # 提取药物的smiles序列
train_drug_smiles = [] # 进一步提取训练集和测试集药物的smiles序列
for d in drugs.values():
drug_smiles.append(d) # 只提取药物对应的smiles
for td in range(len(drug)): # 每批次的药物数量
train_drug_smiles.append(drug_smiles[drug[td]]) # 每批次要获取的药物数量对应的smiles序列
# 读取蛋白质和其sequences序列
proteins = json.load(open("E:\data\kiba\proteins.txt"))
proteins_sequence = [] # 提取蛋白质的序列
train_protein_sequences = [] # 进一步提取训练集蛋白质的sequences序列
for p in proteins.values():
proteins_sequence.append(p) # 只提取蛋白质对应的sequences
for tp in range(len(protein)): # 每批次的蛋白质数量
train_protein_sequences.append(proteins_sequence[protein[td]]) # 每批次要获取的蛋白质数量对应的sequences序列
# 对药物进行label encoding
# 根据smiles序列构成的元素类型数目(64种)进行数值重标设置
CHARISOSMISET = {"#": 29, "%": 30, ")": 31, "(": 1, "+": 32, "-": 33, "/": 34, ".": 2,
"1": 35, "0": 3, "3": 36, "2": 4, "5": 37, "4": 5, "7": 38, "6": 6,
"9": 39, "8": 7, "=": 40, "A": 41, "@": 8, "C": 42, "B": 9, "E": 43,
"D": 10, "G": 44, "F": 11, "I": 45, "H": 12, "K": 46, "M": 47, "L": 13,
"O": 48, "N": 14, "P": 15, "S": 49, "R": 16, "U": 50, "T": 17, "W": 51,
"V": 18, "Y": 52, "[": 53, "Z": 19, "]": 54, "\\": 20, "a": 55, "c": 56,
"b": 21, "e": 57, "d": 22, "g": 58, "f": 23, "i": 59, "h": 24, "m": 60,
"l": 25, "o": 61, "n": 26, "s": 62, "r": 27, "u": 63, "t": 28, "y": 64}
CHARISOSMILEN = 64
drug_label_encoding = []
def label_smiles(smiles): # 药物label encoding函数
D = np.zeros(100) # 药物序列长度设置为100,多切少补0
for i, ch in enumerate(smiles):
if i < 100: # 切片
D[i] = CHARISOSMISET[ch]
return D.tolist()
for l in range(len(train_drug_smiles)):
label_smile = label_smiles(train_drug_smiles[l])
drug_label_encoding.append(label_smile) # 获得药物全部label encoding后转换的数据
# 对蛋白质进行label encoding
# 根据sequence序列构成的元素类型数目(25种)进行数值重标设置
CHARPROTSET = {"A": 1, "C": 2, "B": 3, "E": 4, "D": 5, "G": 6,
"F": 7, "I": 8, "H": 9, "K": 10, "M": 11, "L": 12,
"O": 13, "N": 14, "Q": 15, "P": 16, "S": 17, "R": 18,
"U": 19, "T": 20, "W": 21,
"V": 22, "Y": 23, "X": 24,
"Z": 25}
CHARPROTLEN = 25
protein_label_encoding = []
def label_sequece(sequence): # 蛋白质label encoding函数
P = np.zeros(1000) # 蛋白质序列长度设置为1000,多切少补0
for i, ch in enumerate(sequence):
if i < 1000: # 切片
P[i] = CHARPROTSET[ch]
return P.tolist()
for l in range(len(train_protein_sequences)):
label_protein = label_sequece(train_protein_sequences[l])
protein_label_encoding.append(label_protein) # 获得蛋白质全部label encoding后转换的数据
# 为避免词向量每次嵌入都数值不同设定随机数种子
t.manual_seed(100)
# 对药物进行embedding
drug_label_encodings = t.LongTensor(drug_label_encoding) # 将list转为tensor数据格式
drug_embedding = nn.Embedding(100, 128, padding_idx=0) # 创建药物embedding层
drug_embeddings = drug_embedding(drug_label_encodings) # 嵌入层转换生成128维的词向量,数据维度是[256,100,128]
# 对蛋白质进行embedding
protein_label_encodings = t.LongTensor(protein_label_encoding) # 将list转为tensor数据格式
protein_embedding = nn.Embedding(1000, 128, padding_idx=0) # 创建蛋白质embedding层
protein_embeddings = protein_embedding(protein_label_encodings) # 嵌入层转换生成128维的词向量,数据维度是[256,1000,128]
return drug_embeddings, protein_embeddings, effectives
# 构建Net模块
class Net(nn.Module):
def __init__(self):
super(Net, self).__init__() # 表示Net继承nn.Module
self.cnn1 = nn.Sequential( # 构建Sequential,属于特殊的module,类似于forward前向传播函数,同样的方式调用执行
nn.Conv1d(128, 32, 4, bias=False), # 卷积层,输入通道数为128,输出通道数为32,卷积核为[128,4],包含在Sequential的子module,层层按顺序自动执行
nn.ReLU(inplace=True),
nn.Conv1d(32, 64, 6, bias=False), # 卷积层,输入通道数为32,输出通道数为64,卷积核为[32,6],包含在Sequential的子module,层层按顺序自动执行
nn.ReLU(inplace=True),
nn.Conv1d(64, 96, 8, bias=False), # 卷积层,输入通道数为64,输出通道数为96,卷积核为[64,8],包含在Sequential的子module,层层按顺序自动执行
nn.ReLU(inplace=True),
nn.MaxPool1d(2, 1)
)
self.cnn2 = nn.Sequential( # 构建Sequential,属于特殊的module,类似于forward前向传播函数,同样的方式调用执行
nn.Conv1d(128, 32, 4, bias=False), # 卷积层,输入通道数为128,输出通道数为32,卷积核为[128,4],包含在Sequential的子module,层层按顺序自动执行
nn.ReLU(inplace=True),
nn.Conv1d(32, 64, 8, bias=False), # 卷积层,输入通道数为32,输出通道数为64,卷积核为[32,8],包含在Sequential的子module,层层按顺序自动执行
nn.ReLU(inplace=True),
nn.Conv1d(64, 96, 12, bias=False), # 卷积层,输入通道数为64,输出通道数为96,卷积核为[64,12],包含在Sequential的子module,层层按顺序自动执行
nn.ReLU(inplace=True),
nn.MaxPool1d(2, 1)
)
self.mtp = nn.Sequential( # 构建Sequential,属于特殊的module,类似于forward前向传播函数,同样的方式调用执行
nn.Linear(101952,1024), # 线性连接层,输入通道数为101952=1062*96,输出通道数为1024
nn.Dropout(0.1),
nn.Linear(1024, 512), # 线性连接层,输入通道数为1024,输出通道数为512
nn.Dropout(0.1),
nn.Linear(512, 2) # 线性连接层,输入通道数为512,输出通道数为2
)
def forward(self, drug ,protein): # Net类的前向传播函数
# 维度转换(必要性在于一维卷积的卷积核是(输入通道数,卷积核长),权重参数是(批次,输入通道数,卷积核长))
drug = drug.permute(0, 2, 1)
protein = protein.permute(0, 2, 1)
# CNN卷积操作
drug_cnn = self.cnn1(drug) # 和调用forward一样如此调用cnn1这个Sequential
protein_cnn = self.cnn2(protein) # 和调用forward一样如此调用cnn2这个Sequential
# 卷积完成后再次维度转换为DIT拼接做准备,每批中单个药物的维度是[84,96],每批中单个蛋白质的维度是[978,96]
drug_pool = drug_cnn.permute(0, 2, 1)
protein_pool = protein_cnn.permute(0, 2, 1)
# print(drug_pool.shape)
# print(protein_pool.shape)
# 将训练集和测试集每组对应下标的药物和蛋白质按列进行拼接,每批中单个药物蛋白质结合的维度变成[84+978,96]=[1062,96]
drug_target = t.cat((drug_pool, protein_pool), 1)
# 为全连接数据输入调整tensor的形状,由三维变成二维数据,分别表示[批次大小,每批次数据长度]
drug_target = drug_target.view(256,1062*96)
# 将数据导入全连接层进行二分类任务,亲和和非亲和也就是0和1分类
drug_target_mtp = self.mtp(drug_target)
# 为损失函数计算做准备
drug_target_mtp = t.FloatTensor(drug_target_mtp)
return drug_target_mtp
# 读取训练数据集
train_folds = json.load(open("E:/data/kiba/folds/train_fold_setting.txt"))
# 训练数据集分为5组,每组里面有19709个数据,每个数据值对应亲和矩阵Y中的某一组蛋白质和药物结合对
loaders = t.utils.data.DataLoader(train_folds[0][:1024], batch_size=256, shuffle=False, drop_last=True) # 读取训练数据集数据
# 训练模型
epoch = 3 # 每轮训练次数
model = Net() # 建模
# 计算损失函数值
optimizer = t.optim.SGD(model.parameters(),lr=0.01) # 设置优化器用于更新权重参数,学习率为0.01
for i in range(epoch):
loss_sum = 0
for train_datas in loaders:
# print(train_datas.shape)
# 训练集数据转换
drug_embeddings, protein_embeddings, effectives = datatransform(train_datas)
# 数据输入模型训练
output = model(drug_embeddings, protein_embeddings)
# print(output.shape)
# print(effectives.shape)
# 计算损失函数值
loss = F.nll_loss(output,effectives)
# 每次反向传播前需要梯度清零
optimizer.zero_grad()
# 反向传播
loss.backward(retain_graph=True)
# loss_sum += loss.data.item()
# print(loss_sum)
# 参数权重更新
optimizer.step()
# print(loss_sum)
print("第" + str(i+1) + "轮训练")
# 读取测试集
test_folds = json.load(open("E:/data/kiba/folds/test_fold_setting.txt"))
# 测试数据集只有一组,里面有19709个数据,每个数据值对应亲和矩阵Y中的某一组蛋白质和药物结合对
loaderstest = t.utils.data.DataLoader(test_folds, batch_size=256, shuffle=False, drop_last=True) # 读取训练数据集数据
pre_output = []
real_effectives = []
for test_datas in loaderstest:
# 测试集数据转换
drug_embeddings, protein_embeddings, effectives = datatransform(test_datas)
effectives = effectives.tolist()
# 将每批次真实亲和值输出的数据汇总
real_effectives = real_effectives + effectives
# 测试集数据输入模型训练
output = model(drug_embeddings, protein_embeddings)
output = output.tolist()
# 将每批次预测亲和值输出的数据汇总
for i in range(len(test_datas)):
pre_output.append(output[i].index(max(output[i]))) # 根据大小选取0和1
# 输出正确率
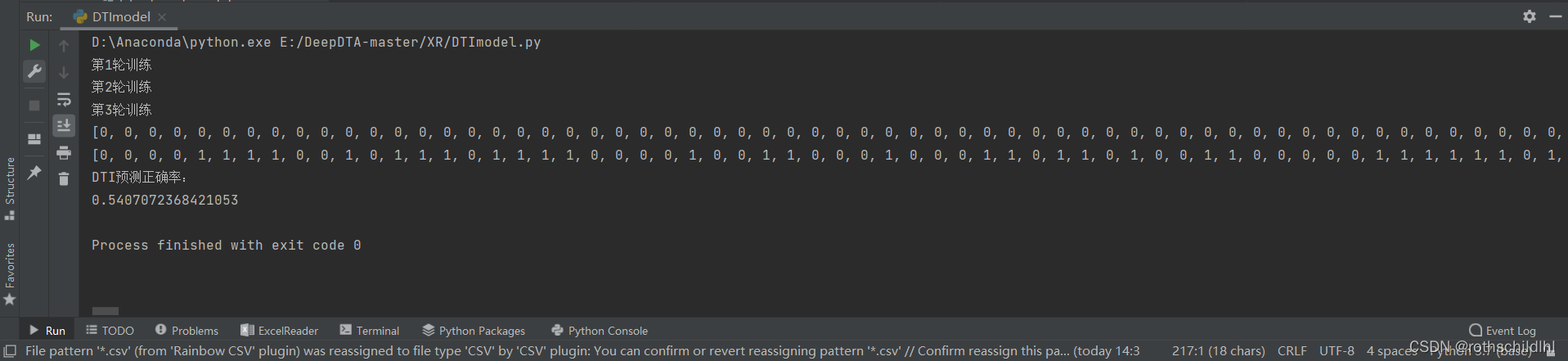
print(pre_output) # 预测输出值
print(real_effectives) # 真实输出值
print("DTI预测正确率:")
print(accuracy_score(real_effectives, pre_output)) # 输出正确率