如何计算一个信号的多尺度熵 Multiscale Entropy
1、多尺度熵,是从样本熵演变过来的
2、样本熵是基于近似熵(ApEn)的一种用于度量时间序列复杂性的改进方法;
SampEn has two advantages over ApEn: data length independence and a relatively trouble-free implementation.
3、首先理解一下什么是近似熵:
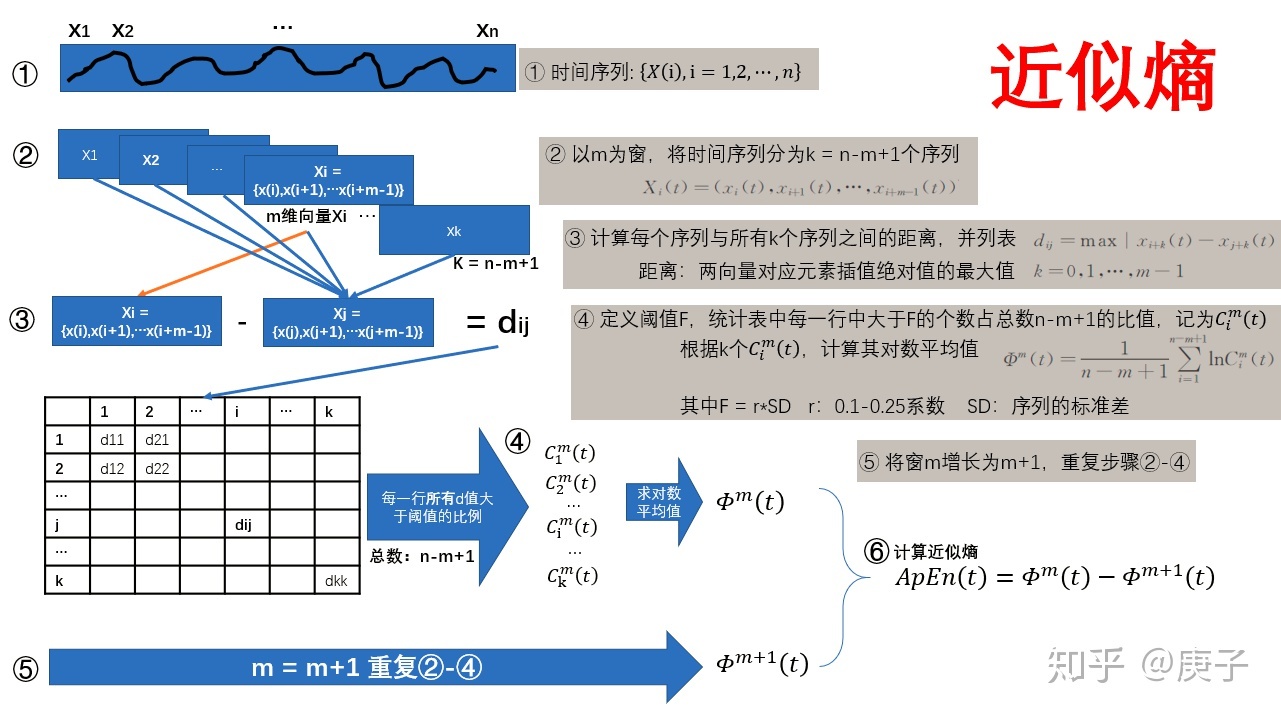
(1)我有个疑问,时间t怎么定义啊;明明k的个片段都用上了,还怎么区分不同的时刻啊。
(2)会不会有一种可能,整个时间序列可以得到一个近似熵的值。也就是一个值。
(3)给一个时间序列后,如果这个时间序列是一维的,你只能得到一个值。
(4)如果这个时间序列是多维的,你可以把某个时刻,这些不同维度的值,组织在一起,计算得到一个熵。然后你就能得到熵随时间的演化了;
4、然后再理解一下,什么是样本熵:
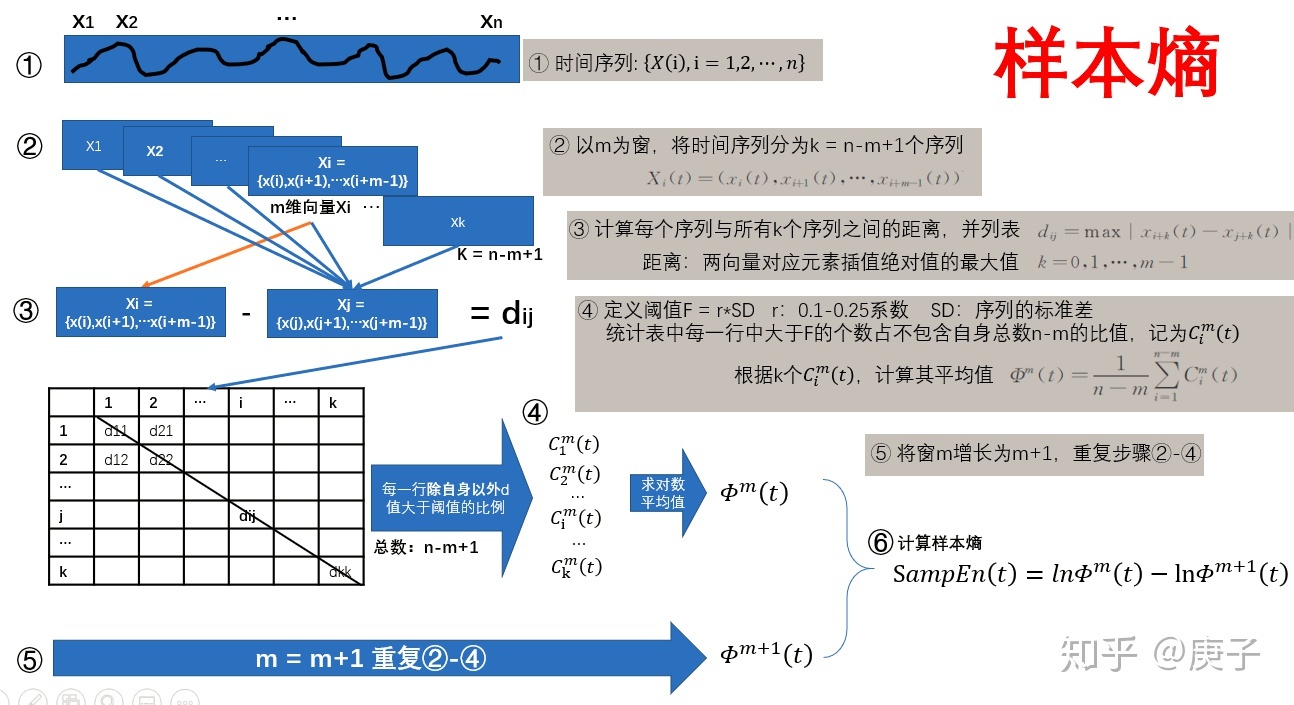
1)近似熵,先取ln再平均。样本熵,平均,再去ln;
2)样本熵,不包含自身。? 近似熵包含了自身;
代码仿真:? wiki
import numpy as np
def sampen(L, m, r):
N = len(L)
B = 0.0
A = 0.0
# Split time series and save all templates of length m
xmi = np.array([L[i : i + m] for i in range(N - m)])
xmj = np.array([L[i : i + m] for i in range(N - m + 1)])
# Save all matches minus the self-match, compute B
B = np.sum([np.sum(np.abs(xmii - xmj).max(axis=1) <= r) - 1 for xmii in xmi])
# Similar for computing A
m += 1
xm = np.array([L[i : i + m] for i in range(N - m + 1)])
A = np.sum([np.sum(np.abs(xmi - xm).max(axis=1) <= r) - 1 for xmi in xm])
# Return SampEn
return -np.log(A / B)
5、还有一个模糊熵;
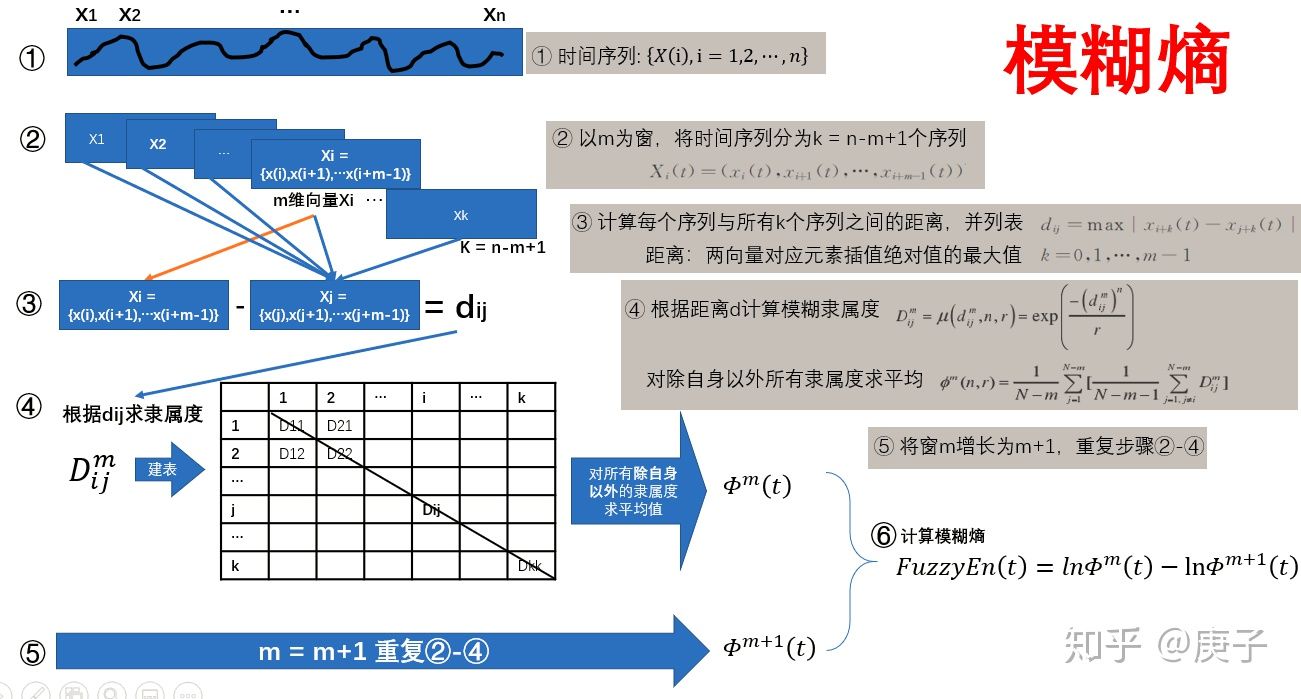
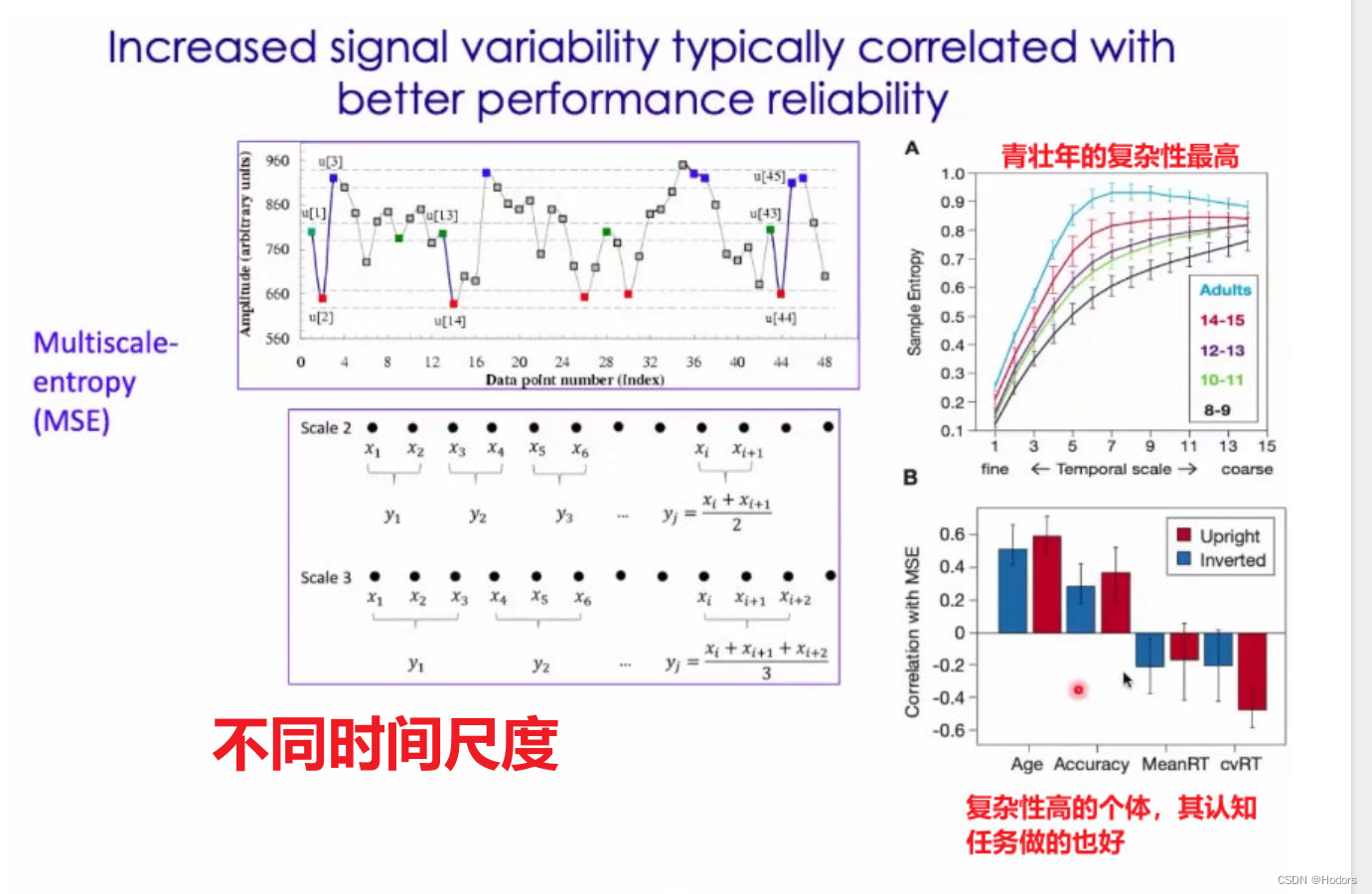
6、最重要的是,多尺度熵。
多尺度熵的基本原理包括对时间序列进行粗粒化或下采样 - 主要是在越来越粗略的时间分辨率下分析时间序列。
样本熵的问题在于它没有很好地考虑到时间序列中可能存在的不同时间尺度。为了计算不同时间尺度下信号的复杂性,Costa等人(2002,2005)提出了多尺度熵。
使用多尺度熵的原因:
1)在单词时间尺度下统计信号的复杂度会比统计整个语音片段的复杂度更加有效。但如果你不知道音频信号代表语音,甚至对语音概念没有任何了解,你就不知道应该运用什么时间尺度以从原始信号中获得更多有用的信息。因此,通过多个时间尺度来分析问题将会得到更多信息。
2)脑电图中,潜在的脑电模式是未知的,因此相关的时间尺度也是未知的。所以,需要通过多尺度样本熵来分析哪个尺度对特定场合下脑电信号的分析更有用.

?
?代码仿真:
% matlab 主程序
clear all;
rand('seed',10)
?
%% generate signals
Sf ?= 1000; ?% Sampling frequency
dur = 30; ?% Time duration
y = colored_noise(Sf,dur,0); ?% white noise
?
%% Plot time course
t = (0:length(y)-1)/Sf; ?% time vector
figure, set(gcf,'Color',[1 1 1]), hold on
?
set(gca,'FontSize',12)
plot(t,y,'-','Color',[0,0,0])
ylim([-3 3])
xlabel('Time (s)')
ylabel('Amp. (a.u.)')
title(['White' ' noise (1/f^{' num2str(0) '})'])
?
%% Plot spectra
[F xf] = spectra(y, Sf, Sf*10, @hann);
figure, set(gcf,'Color',[1 1 1]), hold on
?
set(gca,'FontSize',12)
plot(xf,abs(F),'-','Color',[0,0,0])
% xlim([0 400])
ylim([-0.01 0.06])
xlabel('Frequency (Hz)')
ylabel('Amplitude (a.u.)')
title(['White ' 'noise (1/f^{' num2str(0) '})'])
?
%% Compute MultiScale Entropy
% [mse sf] = MSE_Costa2005(x,nSf,m,r)
%
% x ? - input signal vector (e.g., EEG signal or sound signal)
% nSf - number of scale factors
% m ? - template length (epoch length); Costa used m = 2 throughout?
% r ? - matching threshold; typically chosen to be between 10% and 20% of
% ? ? ? the sample deviation of the time series; when x is z-transformed:
% ? ? ? defined the tolerance as r times the standard deviation
%
% mse - multi-scale entropy
% sf ?- scale factor corresponding to mse
?
[mse,sf] = MSE_Costa2005(y,20,2,std(y)*0.15);
? ??
%% Plot mse
figure, set(gcf,'Color',[1 1 1]), hold on, pp = []; labfull = {};
set(gca,'FontSize',12)
pp = plot(sf,mse,'-','Color',[0,0,0],'LineWidth',2);
labfull = ['White' ' noise (1/f^{' num2str(0) '})'];
ylim([0 3])
xlabel('Scale')
ylabel('SampEn')
title('Multi-scale entropy')
legend(pp,labfull)
legend('boxoff')
----MSE_Costa2005----
function [mse,sf] = MSE_Costa2005(x,nSf,m,r)
% [mse(:,ii) sf] = MSE_Costa2005(y(:,ii),20,2,std(y(:,ii))*0.15);
?
% [mse sf] = MSE_Costa2005(x,nSf,m,r)
%
% x ? - input signal vector (e.g., EEG signal or sound signal)
% nSf - number of scale factors
% m ? - template length (epoch length); Costa used m = 2 throughout?
% r ? - matching threshold; typically chosen to be between 10% and 20% of
% ? ? ? the sample deviation of the time series; when x is z-transformed:
% ? ? ? defined the tolerance as r times the standard deviation
%
% mse - multi-scale entropy
% sf ?- scale factor corresponding to mse
%
% Interpretation: Costa interprets higher values of entropy to reflect more
% information at this scale (less predictable when if random). For 1/f
% pretty constant across scales.
%
% References:
% Costa et al. (2002) Multiscale Entropy Analysis of Complex Physiologic
% ? ?Time Series. PHYSICAL REVIEW LETTERS 89
% Costa et al. (2005) Multiscale entropy analysis of biological signals.
% ? ?PHYSICAL REVIEW E 71, 021906.
%
% Requires: SampleEntropy.m
%
% Description: The script calculates multi-scale entropy using a
% coarse-graining approach.
%
% ---------
%
% ? ?Copyright (C) 2017, B. Herrmann
% ? ?This program is free software: you can redistribute it and/or modify
% ? ?it under the terms of the GNU General Public License as published by
% ? ?the Free Software Foundation, either version 3 of the License, or
% ? ?(at your option) any later version.
%
% ? ?This program is distributed in the hope that it will be useful,
% ? ?but WITHOUT ANY WARRANTY; without even the implied warranty of
% ? ?MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. ?See the
% ? ?GNU General Public License for more details.
%
% ? ?You should have received a copy of the GNU General Public License
% ? ?along with this program. ?If not, see <http://www.gnu.org/licenses/>.
%
% ----------------------------------------------------------------------
% B. Herrmann, Email: herrmann.b@gmail.com, 2017-05-06
?
% pre-allocate mse vector
mse = zeros([1 nSf]);
?
% coarse-grain and calculate sample entropy for each scale factor
for ii = 1 : nSf
?? ?% get filter weights
?? ?f = ones([1 ii]);
?? ?f = f/sum(f);
?
?? ?% get coarse-grained time series (i.e., average data within non-overlapping time windows)
?? ?y = filter(f,1,x);
?? ?y = y(length(f):end);
?? ?y = y(1:length(f):end);
?? ?
?? ?% calculate sample entropy
?? ?mse(ii) = SampleEntropy(y,m,r,0);
end
?
% get sacle factors
sf = 1 : nSf;
----SampleEntropy----
function SampEn = SampleEntropy(x,m,r,sflag)
?
% SampEn = SampleEntropy(x,m,r,sflag)
%
% Obligatory inputs:
%?? ?x - input signal vector (e.g., EEG signal or sound signal)
%
% Optional inputs (defaults):
%?? ?m ? ? = 2; ? % template length (epoch length)
%?? ?r ? ? = 0.2; % matching threshold (default r=.2), when standardized: defined the tolerance as r times the standard deviation
%?? ?sflag = 1; ? % 1 - standardize signal (zero mean, std of one), 0 - no standarization
%
% Output:
%?? ?SampEn - sample entropy estimate (for a sine tone of 1s (Fs=44100) and r=0.2, m=2, the program needed 3.3min)
%
% References:
% Richman JS, Moorman, JR (2000) Physiological time series analysis using approximate?
%?? ? ? entropy and sample entropy. Am J Physiol 278:H2039-H2049
% Abasolo D, Hornero R, Espino P, Alvarez D, Poza J (2006) Entropy analysis of the EEG
%?? ? ? background activity in Alzheimer抯 disease patients. Physioogical Measurement 27:241�253.
% Molina-Pico A, Cuesta-Frau D, Aboy M, Crespo C, Mir�-Mart韓ez P, Oltra-Crespo S (2011) Comparative
%?? ? ? study of approximate entropy and sample entropy robustness to spikes. Artificial Intelligence
% ?? ? in Medicine 53:97�106.
%
% The first reference introduces the method and provides all formulas. But see also these two references,?
% because they depict the formulas much clearer. If you need a standard error estimate have a look at (or the papers):
% http://www.physionet.org/physiotools/sampen/
%
% Description: The program calculates the sample entropy (SampEn) of a given signal. SampEn is the negative?
% logarithm of the conditional probability that two sequences similar for m points remain similar at the next
% point, where self-matches are not included in calculating the probability. Thus, a lower value of SampEn
% also indicates more self-similarity in the time series.
%
% ---------
%
% ? ?Copyright (C) 2012, B. Herrmann
% ? ?This program is free software: you can redistribute it and/or modify
% ? ?it under the terms of the GNU General Public License as published by
% ? ?the Free Software Foundation, either version 3 of the License, or
% ? ?(at your option) any later version.
%
% ? ?This program is distributed in the hope that it will be useful,
% ? ?but WITHOUT ANY WARRANTY; without even the implied warranty of
% ? ?MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. ?See the
% ? ?GNU General Public License for more details.
%
% ? ?You should have received a copy of the GNU General Public License
% ? ?along with this program. ?If not, see <http://www.gnu.org/licenses/>.
%
% -----------------------------------------------------------------------------------------------------
% B. Herrmann, Email: herrmann.b@gmail.com, 2012-01-15
?
% check inputs
SampEn = [];
if nargin < 1 || isempty(x), fprintf('Error: x needs to be defined!\n'), return; end
if ~isvector(x), fprintf('Error: x needs to be a vector!\n'), return; end
if nargin < 2 || isempty(m), m = 2; end
if nargin < 3 || isempty(r), r = 0.2; end
if nargin < 4 || isempty(sflag), sflag = 1; end
?
% force x to be column vector
x = x(:);
N = length(x);
?
?% normalize/standardize x vector
if sflag > 0, x = (x - mean(x)) / std(x); end ?
?
% obtain subsequences of the signal
Xam = zeros(N-m,m+1); Xbm = zeros(N-m,m);
for ii = 1 : N-m ? ? ? ? ? ? ? ? ? % although for N-m+1 subsequences could be extracted for m,
?? ?Xam(ii,:) = x(ii:ii+m); ? ? ? ? ?% in the Richman approach only N-m are considered as this gives the same length for m and m+1
?? ?Xbm(ii,:) = x(ii:ii+m-1);
end
?
% obtain number of matches
B = zeros(1,N-m); A = zeros(1,N-m);
for ii = 1 : N-m
?? ?% number of matches for m
?? ?d = abs(bsxfun(@minus,Xbm((1:N-m)~=ii,:),Xbm(ii,:)));
??? ?B(ii) = sum(max(d,[],2) <= r);
??? ?
?? ?% number of matches for m+1
?? ?d = abs(bsxfun(@minus,Xam((1:N-m)~=ii,:),Xam(ii,:)));
??? ?A(ii) = sum(max(d,[],2) <= r);
end
?
% get probablities for two sequences to match
B ?= 1/(N-m-1) * B; ? ? ? ? ? ? ? ? ?% mean number of matches for each subsequence for m
Bm = 1/(N-m) * sum(B); ? ? ? ? ? ? ? % probability that two sequences will match for m points (mean of matches across subsequences)
A ?= 1/(N-m-1) * A; ? ? ? ? ? ? ? ? ?% mean number of matches for each subsequence for m+1
Am = 1/(N-m) * sum(A); ? ? ? ? ? ? ? % probability that two sequences will match for m+1 points (mean of matches across subsequences)
?
cp = Am/Bm; ? ? ? ? ? ? ? ? ? ? ? ? ?% conditional probability
SampEn = -log(cp); ? ? ? ? ? ? ? ? ? % sample entropy
?
?
参考文献:
1、
?2、
多尺度熵---Understanding Multiscale Entropy_木须耐豆皮的博客-CSDN博客_多尺度样本熵
3、
