《MATLAB 神经网络43个案例分析》:第20章 LIBSVM-FarutoUltimate工具箱及GUI版本介绍与使用
1. 前言
《MATLAB 神经网络43个案例分析》是MATLAB技术论坛(www.matlabsky.com)策划,由王小川老师主导,2013年北京航空航天大学出版社出版的关于MATLAB为工具的一本MATLAB实例教学书籍,是在《MATLAB神经网络30个案例分析》的基础上修改、补充而成的,秉承着“理论讲解—案例分析—应用扩展”这一特色,帮助读者更加直观、生动地学习神经网络。
《MATLAB神经网络43个案例分析》共有43章,内容涵盖常见的神经网络(BP、RBF、SOM、Hopfield、Elman、LVQ、Kohonen、GRNN、NARX等)以及相关智能算法(SVM、决策树、随机森林、极限学习机等)。同时,部分章节也涉及了常见的优化算法(遗传算法、蚁群算法等)与神经网络的结合问题。此外,《MATLAB神经网络43个案例分析》还介绍了MATLAB R2012b中神经网络工具箱的新增功能与特性,如神经网络并行计算、定制神经网络、神经网络高效编程等。
近年来随着人工智能研究的兴起,神经网络这个相关方向也迎来了又一阵研究热潮,由于其在信号处理领域中的不俗表现,神经网络方法也在不断深入应用到语音和图像方向的各种应用当中,本文结合书中案例,对其进行仿真实现,也算是进行一次重新学习,希望可以温故知新,加强并提升自己对神经网络这一方法在各领域中应用的理解与实践。自己正好在多抓鱼上入手了这本书,下面开始进行仿真示例,主要以介绍各章节中源码应用示例为主,本文主要基于MATLAB2015b(32位)平台仿真实现,这是本书第二十章LIBSVM-FarutoUltimate工具箱及GUI版本介绍与使用实例,话不多说,开始!
2. MATLAB 仿真示例
打开MATLAB,点击“主页”,点击“打开”,找到示例文件
选中TutorialTest.m,点击“打开”
TutorialTest.m源码如下:
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
%功能:LIBSVM-FarutoUltimate工具箱及GUI版本介绍与使用
%环境:Win7,Matlab2015b
%Modi: C.S
%时间:2022-06-17
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
%% TutorialTest
%% Matlab神经网络43个案例分析
% LIBSVM-FarutoUltimate工具箱及GUI版本介绍与使用
% by 李洋(faruto)
% http://www.matlabsky.com
% Email:faruto@163.com
% http://weibo.com/faruto
% http://blog.sina.com.cn/faruto
% 2013.01.01
%% 若转载请注明:
% faruto and liyang , LIBSVM-farutoUltimateVersion
% a toolbox with implements for support vector machines based on libsvm, 2011.
% Software available at http://www.matlabsky.com
%
% Chih-Chung Chang and Chih-Jen Lin, LIBSVM : a library for
% support vector machines, 2001. Software available at
% http://www.csie.ntu.edu.tw/~cjlin/libsvm
%%
tic;
close all;
clear;
clc;
format compact;
%% test for scaleForSVM
train_data = [1 12;3 4;7 8]
test_data = [9 10;6 2]
[train_scale,test_scale,ps] = scaleForSVM(train_data,test_data,0,1)
%% test for pcaForSVM
load wine_test
whos train_data
[train_scale,test_scale,ps] = scaleForSVM(train_data,test_data,0,1);
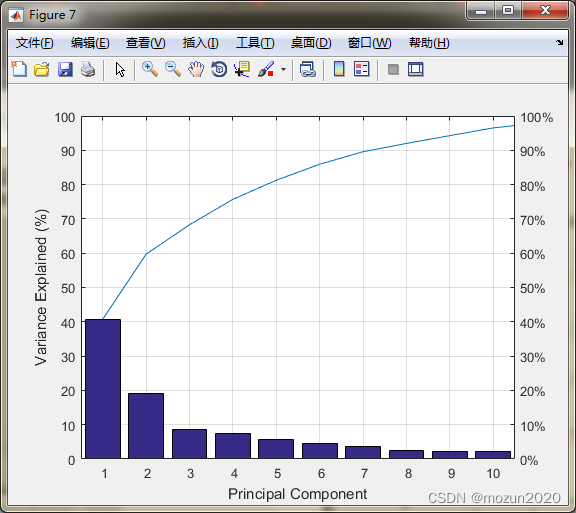
[train_pca,test_pca] = pcaForSVM(train_scale,test_scale,95);
whos train_pca
%% test for SVMcgForClass
load wine_test
[train_scale,test_scale,ps] = scaleForSVM(train_data,test_data,0,1);
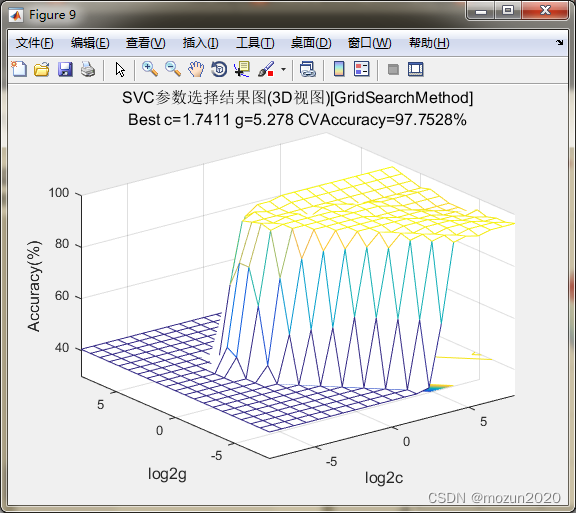
[bestacc,bestc,bestg]=SVMcgForClass(train_data_labels,train_scale,-10,10,-10,10,5,0.5,0.5,4.5)
%% test for psoSVMcgForClass
load wine_test
[train_scale,test_scale,ps] = scaleForSVM(train_data,test_data,0,1);
pso_option.c1 = 1.5;
pso_option.c2 = 1.7;
pso_option.maxgen = 100;
pso_option.sizepop = 20;
pso_option.k = 0.6;
pso_option.wV = 1;
pso_option.wP = 1;
pso_option.v = 5;
pso_option.popcmax = 100;
pso_option.popcmin = 0.1;
pso_option.popgmax = 100;
pso_option.popgmin = 0.1;
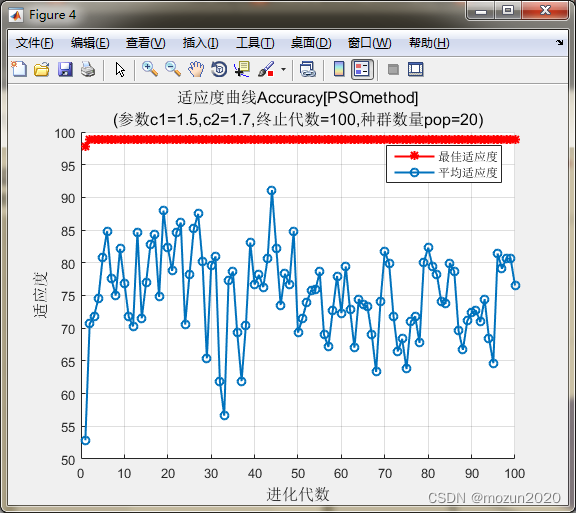
[bestacc,bestc,bestg]=psoSVMcgForClass(train_data_labels,train_scale,pso_option)
%% test for gaSVMcgForClass
load wine_test
[train_scale,test_scale,ps] = scaleForSVM(train_data,test_data,0,1);
ga_option.maxgen = 100;
ga_option.sizepop = 20;
ga_option.cbound = [0,100];
ga_option.gbound = [0,100];
ga_option.v = 10;
ga_option.ggap = 0.9;
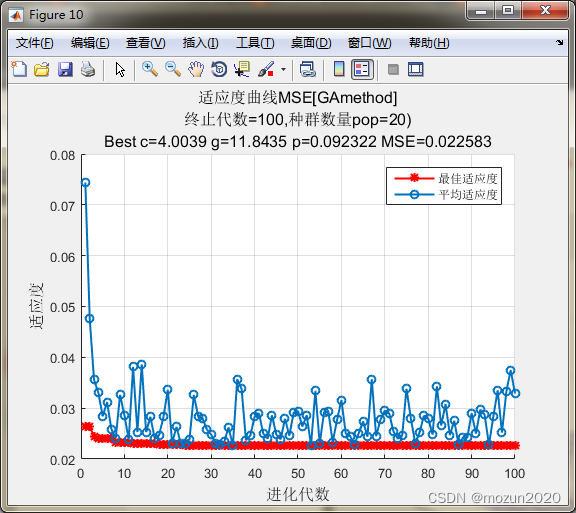
[bestacc,bestc,bestg]=gaSVMcgForClass(train_data_labels,train_scale,ga_option)
%% test for svmplot
load fisheriris;
data = [meas(:,1), meas(:,2)];
groups = ismember(species,'setosa');
[train, test] = crossvalind('holdOut',groups);
dataset = data(train,:);
labels = double(groups(train));
model = svmtrain(labels,dataset,'-c 2 -g 0.1');
svmplot(labels,dataset,model);
%% test for SVC.m
load wine_test;
train_label = train_data_labels;
train_data = train_data;
test_label = test_data_labels;
test_data = test_data;
Method_option.plotOriginal = 0;
Method_option.scale = 1;
Method_option.plotScale = 0;
Method_option.pca = 1;
Method_option.type = 1;
[predict_label,accuracy] = SVC(train_label,train_data,test_label,test_data,Method_option);
%% test for SVR.m
load x123;
train_y = x1(1:17);
train_x = [1:17]';
test_y = x1(18:end,:);
test_x = [18:20]';
Method_option.plotOriginal = 0;
Method_option.xscale = 1;
Method_option.yscale = 1;
Method_option.plotScale = 0;
Method_option.pca = 0;
Method_option.type = 5;
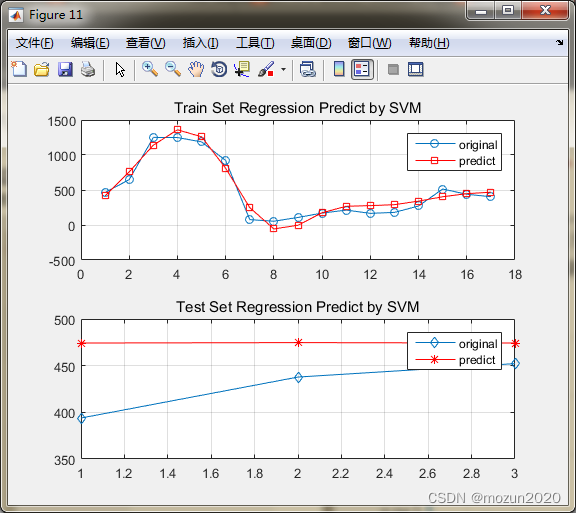
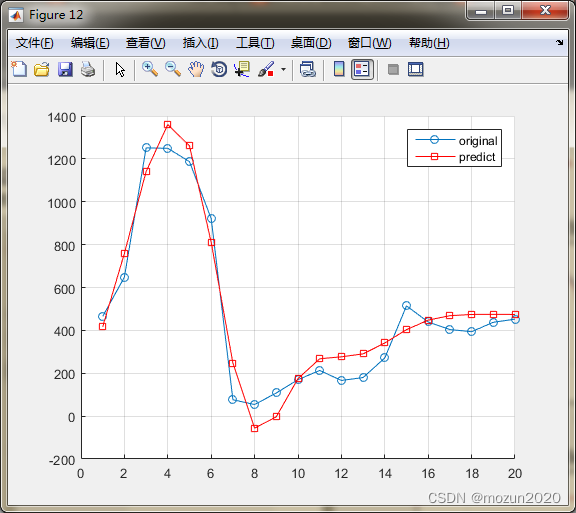
[predict_Y,mse,r] = SVR(train_y,train_x,test_y,test_x,Method_option);
%% test for VF.m
load wine_test
[train_scale,test_scale,ps] = scaleForSVM(train_data,test_data,0,1);
model = svmtrain(train_data_labels,train_scale,'-c 2 -g 0.1');
[pre,acc] = svmpredict(train_data_labels,train_scale,model);
[score,str] = VF(train_data_labels,pre,1)
[score,str] = VF(train_data_labels,pre,2)
[score,str] = VF(train_data_labels,pre,3)
[score,str] = VF(train_data_labels,pre,4)
[score,str] = VF(train_data_labels,pre,5)
%% test for ClassResult.m
load heart_scale.mat;
data = heart_scale_inst;
label = heart_scale_label;
model = svmtrain(label,data);
type = 1;
CR = ClassResult(label, data, model, type)
%% test for plotSVMroc
load wine_test
[train_data,test_data] = scaleForSVM(train_data,test_data,0,1);
model = svmtrain(train_data_labels,train_data,'-c 0.01 -g 0.01 -b 1');
[pre,acc,dec] = svmpredict(train_data_labels,train_data,model,'-b 1');
plotSVMroc(train_data_labels,dec,3)
%%
toc;
添加完毕,点击“运行”,开始仿真,输出仿真结果如下:
train_data =
1 12
3 4
7 8
test_data =
9 10
6 2
train_scale =
0 1.0000
0.2500 0.2000
0.7500 0.6000
test_scale =
1.0000 0.8000
0.6250 0
ps =
name: 'mapminmax'
xrows: 2
xmax: [2x1 double]
xmin: [2x1 double]
xrange: [2x1 double]
yrows: 2
ymax: 1
ymin: 0
yrange: 1
no_change: 0
gain: [2x1 double]
xoffset: [2x1 double]
Name Size Bytes Class Attributes
train_data 89x13 9256 double
Name Size Bytes Class Attributes
train_pca 89x10 7120 double
bestacc =
98.8764
bestc =
2
bestg =
2.8284
bestacc =
98.8764
bestc =
5.1197
bestg =
4.3773
bestacc =
98.8764
bestc =
2.7023
bestg =
3.6726
Mean squared error = 2.71781 (regression)
Squared correlation coefficient = -1.#IND (regression)
bestCVaccuracy =
97.7528
bestc =
1.7411
bestg =
5.2780
Accuracy = 100% (89/89) (classification)
Accuracy = 97.7528% (87/89) (classification)
accuracy =
100.0000 97.7528
bestCVmse =
0.0226
bestc =
4.0039
bestg =
11.8435
bestp =
0.0923
Mean squared error = 0.00654143 (regression)
Squared correlation coefficient = 0.945718 (regression)
Mean squared error = 0.00193148 (regression)
Squared correlation coefficient = 0.0702999 (regression)
mse =
0.0065 0.0019
r =
0.9457 0.0703
Accuracy = 96.6292% (86/89) (classification)
score =
96.6292
str =
Accuracy = 96.6292% (86/89) [Accuracy = #true / #total]
score =
96.7742
str =
Precision = 96.7742% (30/31) [Precision = true_positive / (true_positive + false_positive)]
score =
100
str =
Recall = 100% (30/30) [Recall = true_positive / (true_positive + false_negative)]
score =
98.3607
str =
F-score = 98.3607% [F-score = 2 * Precision * Recall / (Precision + Recall)]
score =
50
str =
BAC = 50% [BAC (Ballanced ACcuracy) = (Sensitivity + Specificity) / 2]
Accuracy = 86.6667% (234/270) (classification)
===some info of the data set===
#class is 2
类别标签为 1 -1
支持向量数目 132,所占训练集样本数目比例 48.8889% (132/270)
===各种分类准确率===
整体分类准确率 = 86.6667% (234/270)
第 1 类分类准确率 = 80.8333% (97/120)
第 -1 类分类准确率 = 91.3333% (137/150)
CR =
accuracy: [0.8667 0.8083 0.9133]
SVlocation: [132x1 double]
b: -0.4245
w: [132x1 double]
alpha: [132x1 double]
Accuracy = 95.5056% (85/89) (classification)
时间已过 29.267572 秒。
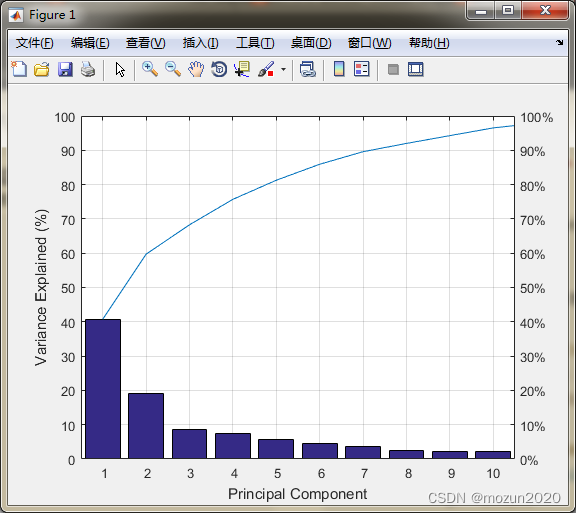
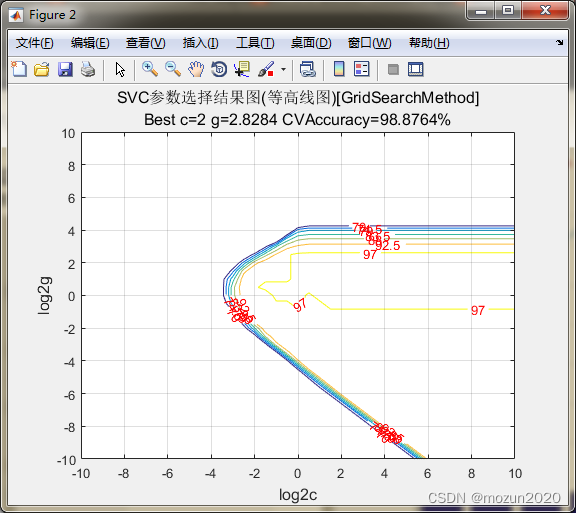
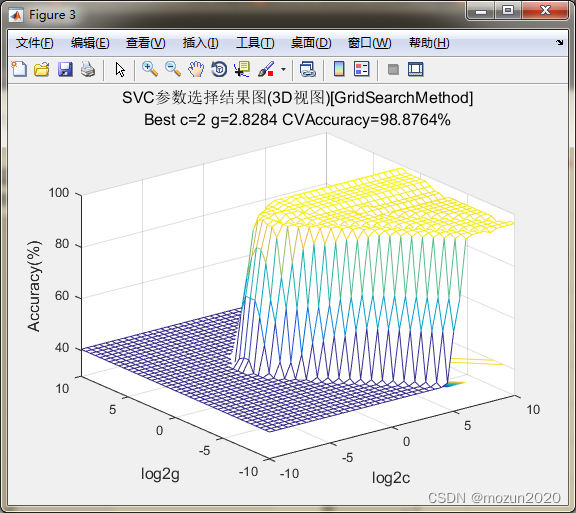





3. 小结
libsvm工具箱的应用还是很多的,前几章的仿真就是也需要这个工具箱中SVM相关的函数需要调用才一直没跑通,后面将工具箱中的路径包含到育英路径中去才完成仿真,工具箱里面的应用还是相对较多的,作为仿真来说也比较适合。对本章内容感兴趣或者想充分学习了解的,建议去研习书中第二十章节的内容。后期会对其中一些知识点在自己理解的基础上进行补充,欢迎大家一起学习交流。