文章大纲
简介
正则化

标准化(standardization)
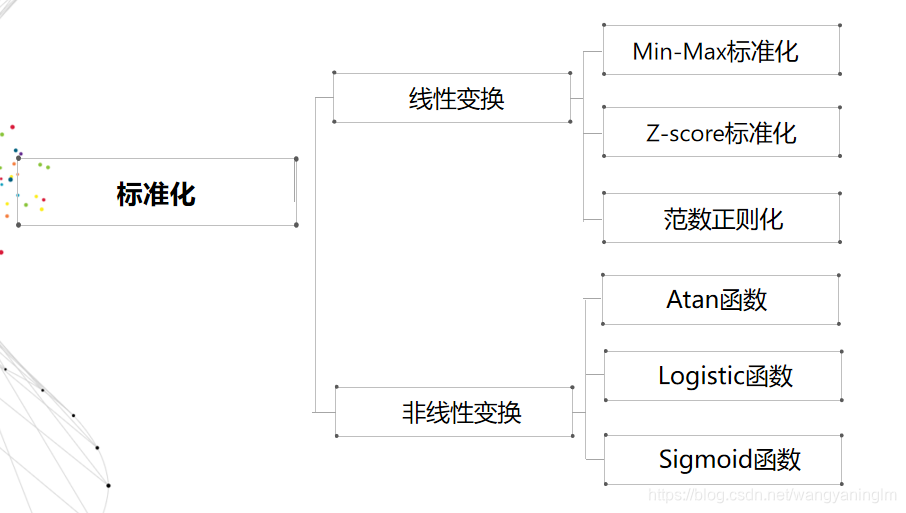
数据标准化是将数据按比例缩放,使其落入到一个小的区间内,标准化后的数据可正可负,但是一般绝对值不会太大,一般是z-score标准化方法:减去期望后除以标准差。
x ? = x ? u σ x^\ast=\frac{x-u}\sigma x?=σx?u?
特点:
对不同特征维度的伸缩变换的目的是使其不同度量之间的特征具有可比性,同时不改变原始数据的分布。
好处:
- 不改变原始数据的分布,保持各个特征维度对目标函数的影响权重
- 对目标函数的影响体现在几何分布上
- 在已有样本足够多的情况下比较稳定,适合现代嘈杂大数据场景
归一化
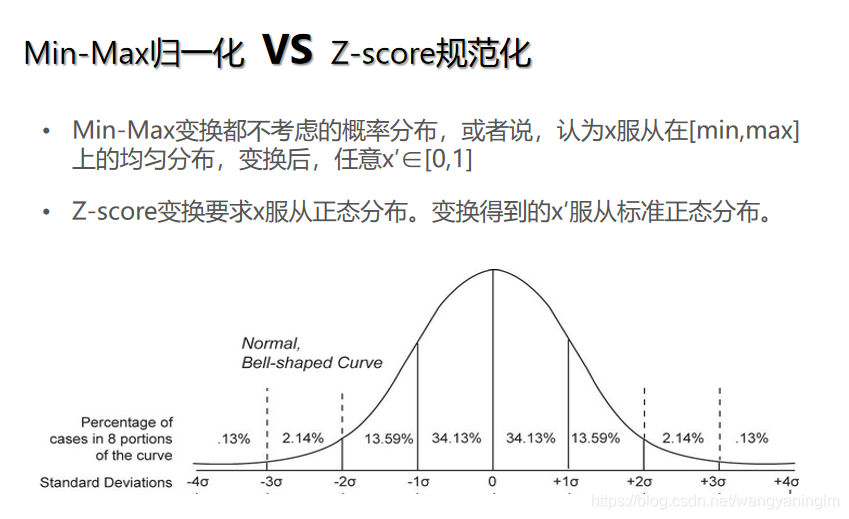
把数值放缩到0到1的小区间中(归到数字信号处理范畴之内),一般方法是最小最大规范的方法:min-max normalization
x ? = x ? m i n m a x ? m i n x^\ast=\frac{x-min}{max-min} x?=max?minx?min?
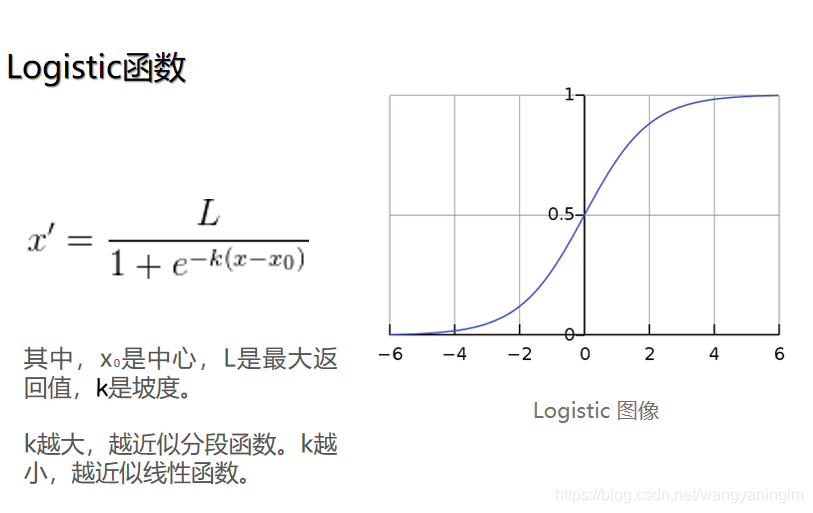
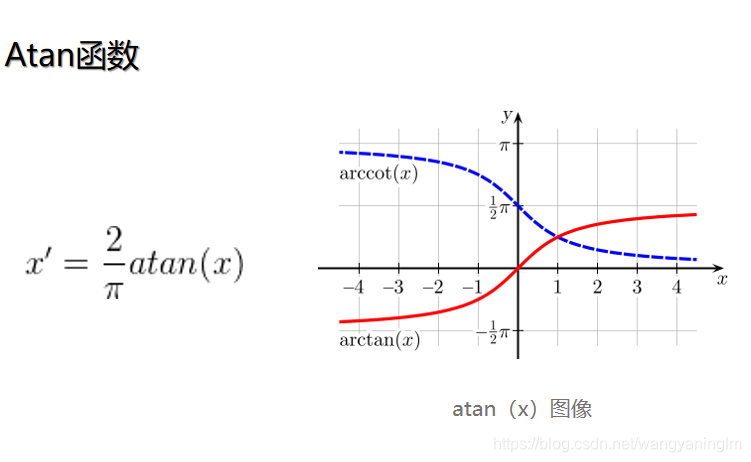
上面min-max normalization是线性归一化,还有非线性归一化,通过一些数学函数,将原始值进行映射。该方法包括log、指数、反正切等。需要根据数据分布的情况,决定非线性函数的曲线。
spark 中的正则化
Normalizer
标准化文档:
- http://spark.apache.org/docs/latest/api/scala/org/apache/spark/ml/feature/Normalizer.html
标准化源代码:
- https://github.com/apache/spark/blob/v3.1.2/mllib/src/main/scala/org/apache/spark/ml/feature/Normalizer.scala
文档中就这么一句话:
Normalize a vector to have unit norm using the given p-norm.
使用给定的p-范数规范化向量,使其具有单位范数。
源代码
package org.apache.spark.ml.feature
import org.apache.spark.annotation.Since
import org.apache.spark.ml.UnaryTransformer
import org.apache.spark.ml.attribute.AttributeGroup
import org.apache.spark.ml.linalg.{Vector, VectorUDT}
import org.apache.spark.ml.param.{DoubleParam, ParamValidators}
import org.apache.spark.ml.util._
import org.apache.spark.mllib.feature
import org.apache.spark.mllib.linalg.{Vectors => OldVectors}
import org.apache.spark.sql.types._
/**
* Normalize a vector to have unit norm using the given p-norm.
*/
@Since("1.4.0")
class Normalizer @Since("1.4.0") (@Since("1.4.0") override val uid: String)
extends UnaryTransformer[Vector, Vector, Normalizer] with DefaultParamsWritable {
@Since("1.4.0")
def this() = this(Identifiable.randomUID("normalizer"))
/**
* Normalization in L^p^ space. Must be greater than equal to 1.
* (default: p = 2)
* @group param
*/
@Since("1.4.0")
val p = new DoubleParam(this, "p", "the p norm value", ParamValidators.gtEq(1))
setDefault(p -> 2.0)
/** @group getParam */
@Since("1.4.0")
def getP: Double = $(p)
/** @group setParam */
@Since("1.4.0")
def setP(value: Double): this.type = set(p, value)
override protected def createTransformFunc: Vector => Vector = {
val normalizer = new feature.Normalizer($(p))
vector => normalizer.transform(OldVectors.fromML(vector)).asML
}
override protected def validateInputType(inputType: DataType): Unit = {
require(inputType.isInstanceOf[VectorUDT],
s"Input type must be ${(new VectorUDT).catalogString} but got ${inputType.catalogString}.")
}
override protected def outputDataType: DataType = new VectorUDT()
@Since("1.4.0")
override def transformSchema(schema: StructType): StructType = {
var outputSchema = super.transformSchema(schema)
if ($(inputCol).nonEmpty && $(outputCol).nonEmpty) {
val size = AttributeGroup.fromStructField(schema($(inputCol))).size
if (size >= 0) {
outputSchema = SchemaUtils.updateAttributeGroupSize(outputSchema,
$(outputCol), size)
}
}
outputSchema
}
@Since("3.0.0")
override def toString: String = {
s"Normalizer: uid=$uid, p=${$(p)}"
}
}
@Since("1.6.0")
object Normalizer extends DefaultParamsReadable[Normalizer] {
@Since("1.6.0")
override def load(path: String): Normalizer = super.load(path)
}
spark 中的标准化
Standardizes
Standardizes features by removing the mean and scaling to unit variance using column summary statistics on the samples in the training set.
The “unit std” is computed using the corrected sample standard deviation, which is computed as the square root of the unbiased sample variance.
代码:
- https://github.com/apache/spark/blob/v3.1.2/mllib/src/main/scala/org/apache/spark/ml/feature/StandardScaler.scala
源代码
package org.apache.spark.ml.feature
import org.apache.hadoop.fs.Path
import org.apache.spark.annotation.Since
import org.apache.spark.ml._
import org.apache.spark.ml.linalg._
import org.apache.spark.ml.param._
import org.apache.spark.ml.param.shared._
import org.apache.spark.ml.stat.Summarizer
import org.apache.spark.ml.util._
import org.apache.spark.mllib.util.MLUtils
import org.apache.spark.sql._
import org.apache.spark.sql.functions._
import org.apache.spark.sql.types.{StructField, StructType}
/**
* Params for [[StandardScaler]] and [[StandardScalerModel]].
*/
private[feature] trait StandardScalerParams extends Params with HasInputCol with HasOutputCol {
/**
* Whether to center the data with mean before scaling.
* It will build a dense output, so take care when applying to sparse input.
* Default: false
* @group param
*/
val withMean: BooleanParam = new BooleanParam(this, "withMean",
"Whether to center data with mean")
/** @group getParam */
def getWithMean: Boolean = $(withMean)
/**
* Whether to scale the data to unit standard deviation.
* Default: true
* @group param
*/
val withStd: BooleanParam = new BooleanParam(this, "withStd",
"Whether to scale the data to unit standard deviation")
/** @group getParam */
def getWithStd: Boolean = $(withStd)
/** Validates and transforms the input schema. */
protected def validateAndTransformSchema(schema: StructType): StructType = {
SchemaUtils.checkColumnType(schema, $(inputCol), new VectorUDT)
require(!schema.fieldNames.contains($(outputCol)),
s"Output column ${$(outputCol)} already exists.")
val outputFields = schema.fields :+ StructField($(outputCol), new VectorUDT, false)
StructType(outputFields)
}
setDefault(withMean -> false, withStd -> true)
}
/**
* Standardizes features by removing the mean and scaling to unit variance using column summary
* statistics on the samples in the training set.
*
* The "unit std" is computed using the
* <a href="https://en.wikipedia.org/wiki/Standard_deviation#Corrected_sample_standard_deviation">
* corrected sample standard deviation</a>,
* which is computed as the square root of the unbiased sample variance.
*/
@Since("1.2.0")
class StandardScaler @Since("1.4.0") (
@Since("1.4.0") override val uid: String)
extends Estimator[StandardScalerModel] with StandardScalerParams with DefaultParamsWritable {
@Since("1.2.0")
def this() = this(Identifiable.randomUID("stdScal"))
/** @group setParam */
@Since("1.2.0")
def setInputCol(value: String): this.type = set(inputCol, value)
/** @group setParam */
@Since("1.2.0")
def setOutputCol(value: String): this.type = set(outputCol, value)
/** @group setParam */
@Since("1.4.0")
def setWithMean(value: Boolean): this.type = set(withMean, value)
/** @group setParam */
@Since("1.4.0")
def setWithStd(value: Boolean): this.type = set(withStd, value)
@Since("2.0.0")
override def fit(dataset: Dataset[_]): StandardScalerModel = {
transformSchema(dataset.schema, logging = true)
val Row(mean: Vector, std: Vector) = dataset
.select(Summarizer.metrics("mean", "std").summary(col($(inputCol))).as("summary"))
.select("summary.mean", "summary.std")
.first()
copyValues(new StandardScalerModel(uid, std.compressed, mean.compressed).setParent(this))
}
@Since("1.4.0")
override def transformSchema(schema: StructType): StructType = {
validateAndTransformSchema(schema)
}
@Since("1.4.1")
override def copy(extra: ParamMap): StandardScaler = defaultCopy(extra)
}
@Since("1.6.0")
object StandardScaler extends DefaultParamsReadable[StandardScaler] {
@Since("1.6.0")
override def load(path: String): StandardScaler = super.load(path)
}
/**
* Model fitted by [[StandardScaler]].
*
* @param std Standard deviation of the StandardScalerModel
* @param mean Mean of the StandardScalerModel
*/
@Since("1.2.0")
class StandardScalerModel private[ml] (
@Since("1.4.0") override val uid: String,
@Since("2.0.0") val std: Vector,
@Since("2.0.0") val mean: Vector)
extends Model[StandardScalerModel] with StandardScalerParams with MLWritable {
import StandardScalerModel._
/** @group setParam */
@Since("1.2.0")
def setInputCol(value: String): this.type = set(inputCol, value)
/** @group setParam */
@Since("1.2.0")
def setOutputCol(value: String): this.type = set(outputCol, value)
@Since("2.0.0")
override def transform(dataset: Dataset[_]): DataFrame = {
val outputSchema = transformSchema(dataset.schema, logging = true)
val shift = if ($(withMean)) mean.toArray else Array.emptyDoubleArray
val scale = if ($(withStd)) {
std.toArray.map { v => if (v == 0) 0.0 else 1.0 / v }
} else Array.emptyDoubleArray
val func = getTransformFunc(shift, scale, $(withMean), $(withStd))
val transformer = udf(func)
dataset.withColumn($(outputCol), transformer(col($(inputCol))),
outputSchema($(outputCol)).metadata)
}
@Since("1.4.0")
override def transformSchema(schema: StructType): StructType = {
var outputSchema = validateAndTransformSchema(schema)
if ($(outputCol).nonEmpty) {
outputSchema = SchemaUtils.updateAttributeGroupSize(outputSchema,
$(outputCol), mean.size)
}
outputSchema
}
@Since("1.4.1")
override def copy(extra: ParamMap): StandardScalerModel = {
val copied = new StandardScalerModel(uid, std, mean)
copyValues(copied, extra).setParent(parent)
}
@Since("1.6.0")
override def write: MLWriter = new StandardScalerModelWriter(this)
@Since("3.0.0")
override def toString: String = {
s"StandardScalerModel: uid=$uid, numFeatures=${mean.size}, withMean=${$(withMean)}, " +
s"withStd=${$(withStd)}"
}
}
@Since("1.6.0")
object StandardScalerModel extends MLReadable[StandardScalerModel] {
private[StandardScalerModel]
class StandardScalerModelWriter(instance: StandardScalerModel) extends MLWriter {
private case class Data(std: Vector, mean: Vector)
override protected def saveImpl(path: String): Unit = {
DefaultParamsWriter.saveMetadata(instance, path, sc)
val data = Data(instance.std, instance.mean)
val dataPath = new Path(path, "data").toString
sparkSession.createDataFrame(Seq(data)).repartition(1).write.parquet(dataPath)
}
}
private class StandardScalerModelReader extends MLReader[StandardScalerModel] {
private val className = classOf[StandardScalerModel].getName
override def load(path: String): StandardScalerModel = {
val metadata = DefaultParamsReader.loadMetadata(path, sc, className)
val dataPath = new Path(path, "data").toString
val data = sparkSession.read.parquet(dataPath)
val Row(std: Vector, mean: Vector) = MLUtils.convertVectorColumnsToML(data, "std", "mean")
.select("std", "mean")
.head()
val model = new StandardScalerModel(metadata.uid, std, mean)
metadata.getAndSetParams(model)
model
}
}
@Since("1.6.0")
override def read: MLReader[StandardScalerModel] = new StandardScalerModelReader
@Since("1.6.0")
override def load(path: String): StandardScalerModel = super.load(path)
private[spark] def transformWithBoth(
shift: Array[Double],
scale: Array[Double],
values: Array[Double]): Array[Double] = {
var i = 0
while (i < values.length) {
values(i) = (values(i) - shift(i)) * scale(i)
i += 1
}
values
}
private[spark] def transformWithShift(
shift: Array[Double],
values: Array[Double]): Array[Double] = {
var i = 0
while (i < values.length) {
values(i) -= shift(i)
i += 1
}
values
}
private[spark] def transformDenseWithScale(
scale: Array[Double],
values: Array[Double]): Array[Double] = {
var i = 0
while (i < values.length) {
values(i) *= scale(i)
i += 1
}
values
}
private[spark] def transformSparseWithScale(
scale: Array[Double],
indices: Array[Int],
values: Array[Double]): Array[Double] = {
var i = 0
while (i < values.length) {
values(i) *= scale(indices(i))
i += 1
}
values
}
private[spark] def getTransformFunc(
shift: Array[Double],
scale: Array[Double],
withShift: Boolean,
withScale: Boolean): Vector => Vector = {
(withShift, withScale) match {
case (true, true) =>
vector: Vector =>
val values = vector match {
case d: DenseVector => d.values.clone()
case v: Vector => v.toArray
}
val newValues = transformWithBoth(shift, scale, values)
Vectors.dense(newValues)
case (true, false) =>
vector: Vector =>
val values = vector match {
case d: DenseVector => d.values.clone()
case v: Vector => v.toArray
}
val newValues = transformWithShift(shift, values)
Vectors.dense(newValues)
case (false, true) =>
vector: Vector =>
vector match {
case DenseVector(values) =>
val newValues = transformDenseWithScale(scale, values.clone())
Vectors.dense(newValues)
case SparseVector(size, indices, values) =>
val newValues = transformSparseWithScale(scale, indices, values.clone())
Vectors.sparse(size, indices, newValues)
case v =>
throw new IllegalArgumentException(s"Unknown vector type ${v.getClass}.")
}
case (false, false) =>
vector: Vector => vector
}
}
}
spark 中的归一化
MaxAbsScaler
- http://spark.apache.org/docs/latest/api/scala/org/apache/spark/ml/feature/MaxAbsScaler.html
MinMaxScaler
- http://spark.apache.org/docs/latest/api/scala/org/apache/spark/ml/feature/MinMaxScaler.html
Rescale each feature individually to a common range [min, max] linearly using column summary
statistics, which is also known as min-max normalization or Rescaling. The rescaled value for
feature E is calculated as:
R
e
s
c
a
l
e
d
(
e
i
)
=
e
i
?
E
m
i
n
E
m
a
x
?
E
m
i
n
?
(
m
a
x
?
m
i
n
)
+
m
i
n
Rescaled(e_i) = \frac{e_i - E_{min}}{E_{max} - E_{min}} * (max - min) + min
Rescaled(ei?)=Emax??Emin?ei??Emin???(max?min)+min
For the case (E_{max} == E_{min}), (Rescaled(e_i) = 0.5 * (max + min)).
note :
Since zero values will probably be transformed to non-zero values, output of the
transformer will be DenseVector even for sparse input.
核心代码:主要就是计算 最大最小值
override def fit(dataset: Dataset[_]): MinMaxScalerModel = {
transformSchema(dataset.schema, logging = true)
val Row(max: Vector, min: Vector) = dataset
.select(Summarizer.metrics("max", "min").summary(col($(inputCol))).as("summary"))
.select("summary.max", "summary.min")
.first()
copyValues(new MinMaxScalerModel(uid, min.compressed, max.compressed).setParent(this))
}
参考文献
spark 中的 特征相关内容处理的文档
- http://spark.apache.org/docs/latest/api/scala/org/apache/spark/ml/feature/index.html
概念简介
- https://blog.csdn.net/u014381464/article/details/81101551
参考:
- https://segmentfault.com/a/1190000014042959
- https://www.cnblogs.com/nucdy/p/7994542.html
- https://blog.csdn.net/weixin_34117522/article/details/88875270
- https://blog.csdn.net/xuejianbest/article/details/85779029