一、简介
支持向量机(Support Vector Machine)是Cortes和Vapnik于1995年首先提出的,它在解决小样本、非线性及高维模式识别中表现出许多特有的优势,并能够推广应用到函数拟合等其他机器学习问题中。
1 数学部分
1.1 二维空间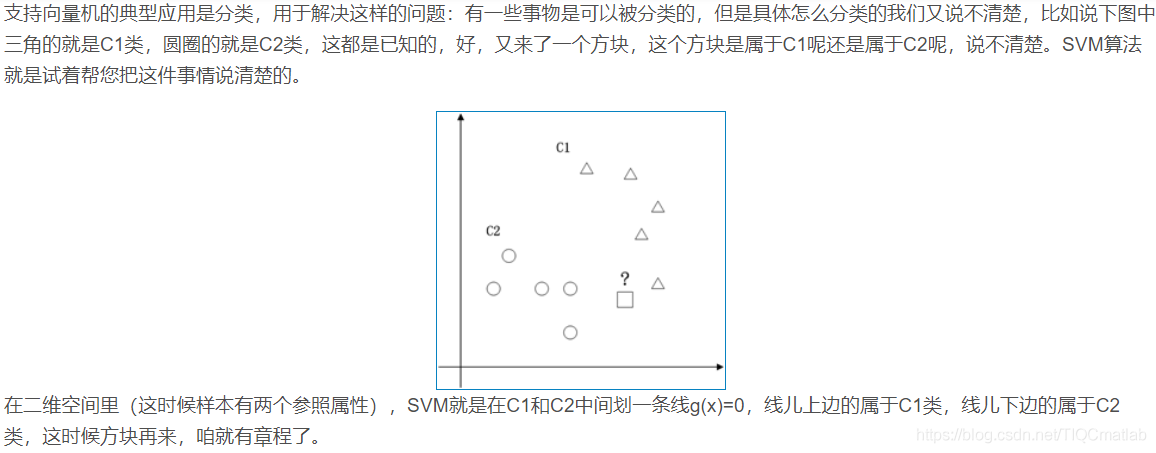
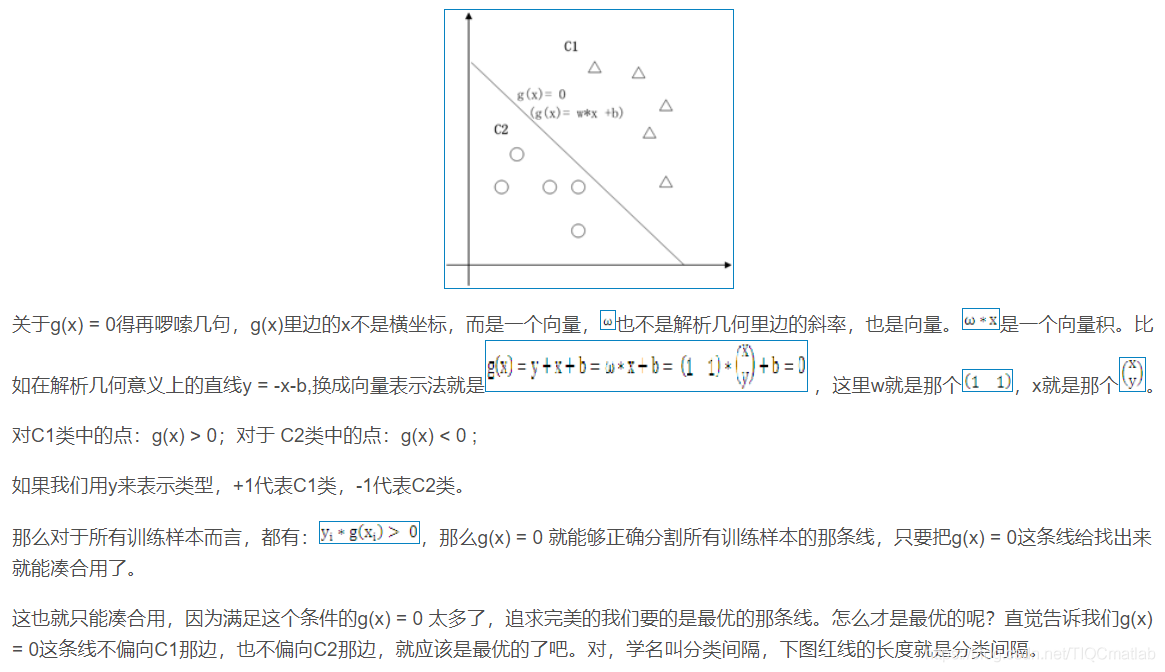
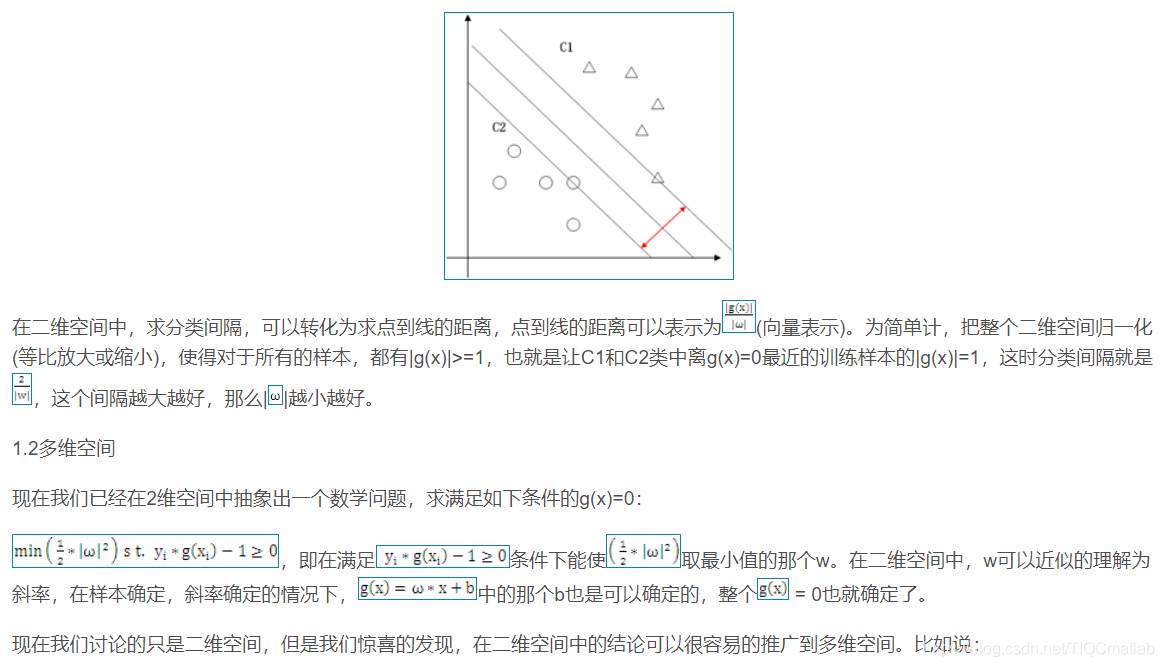
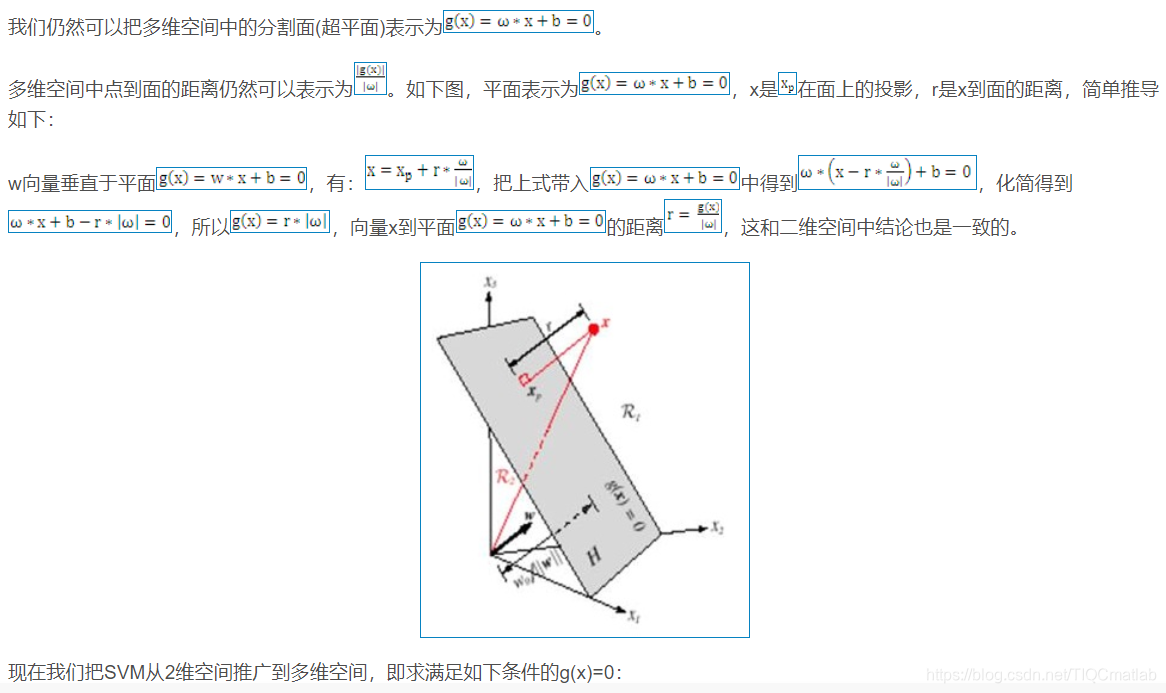


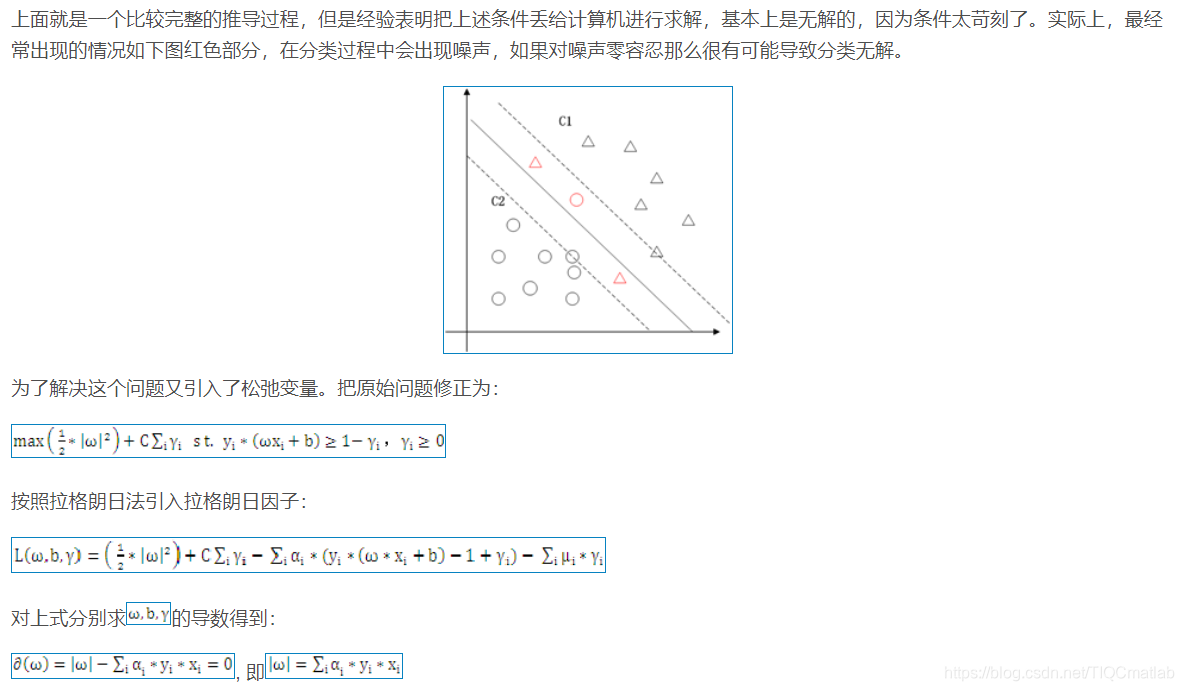


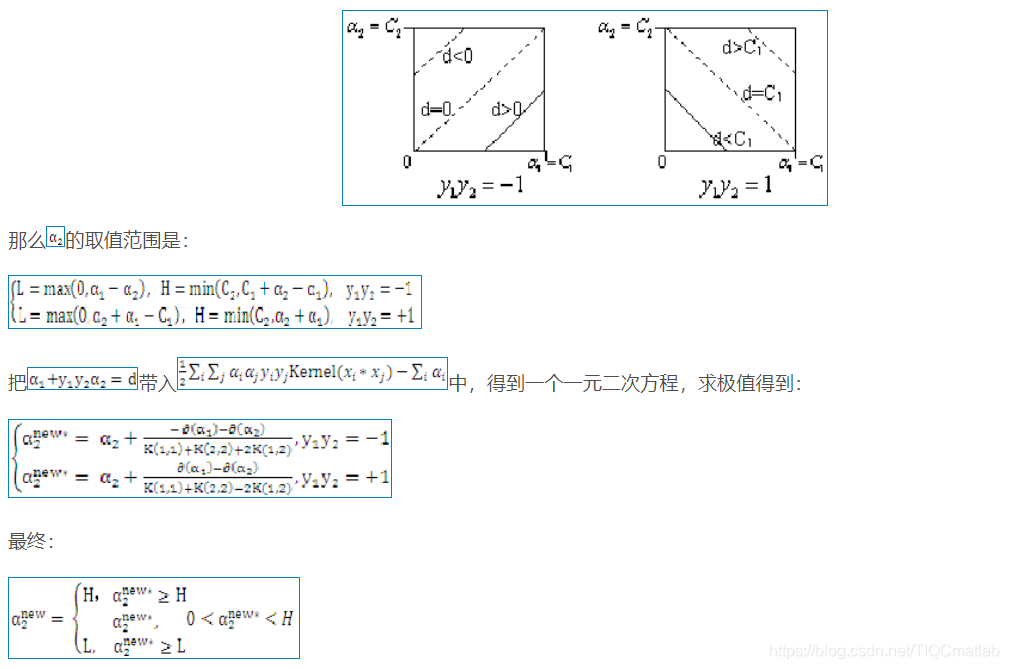
2 算法部分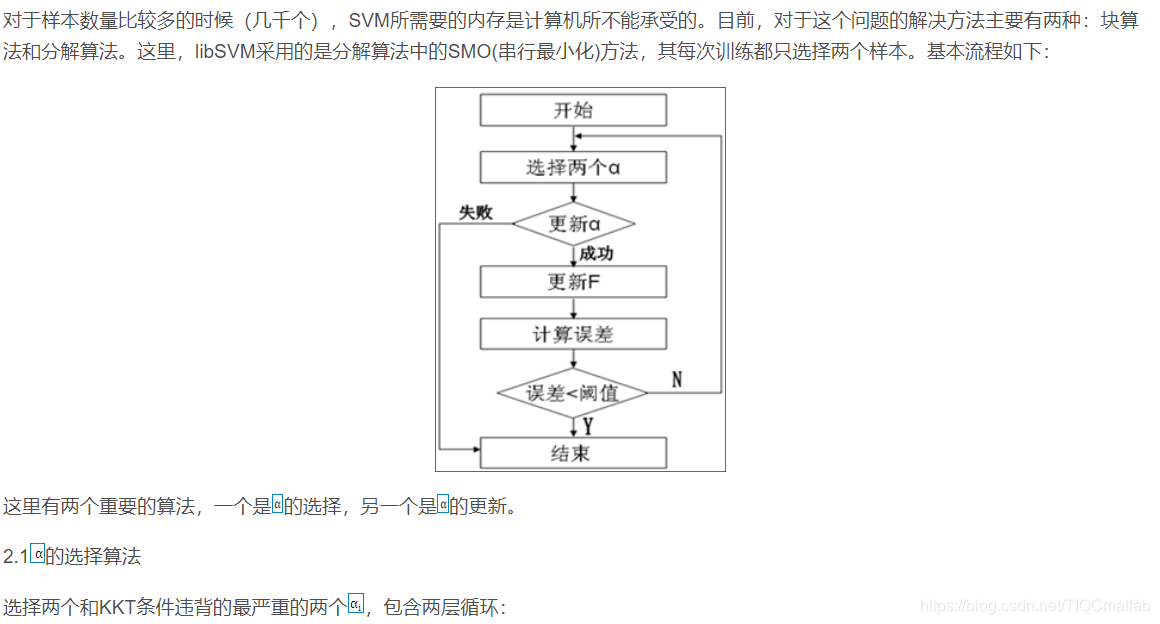


二、源代码
function [model,H] = lssvmMATLAB(model)
% Only for intern LS-SVMlab use;
%
% MATLAB implementation of the LS-SVM algorithm. This is slower
% than the C-mex implementation, but it is more reliable and flexible;
%
%
% This implementation is quite straightforward, based on MATLAB's
% backslash matrix division (or PCG if available) and total kernel
% matrix construction. It has some extensions towards advanced
% techniques, especially applicable on small datasets (weighed
% LS-SVM, gamma-per-datapoint)
% Copyright (c) 2002, KULeuven-ESAT-SCD, License & help @ http://www.esat.kuleuven.ac.be/sista/lssvmlab
%fprintf('~');
%
% is it weighted LS-SVM ?
%
weighted = (length(model.gam)>model.y_dim);
if and(weighted,length(model.gam)~=model.nb_data),
warning('not enough gamma''s for Weighted LS-SVMs, simple LS-SVM applied');
weighted=0;
end
% computation omega and H
omega = kernel_matrix(model.xtrain(model.selector, 1:model.x_dim), ...
model.kernel_type, model.kernel_pars);
% initiate alpha and b
model.b = zeros(1,model.y_dim);
model.alpha = zeros(model.nb_data,model.y_dim);
for i=1:model.y_dim,
H = omega;
model.selector=~isnan(model.ytrain(:,i));
nb_data=sum(model.selector);
if size(model.gam,2)==model.nb_data,
try invgam = model.gam(i,:).^-1; catch, invgam = model.gam(1,:).^-1;end
for t=1:model.nb_data, H(t,t) = H(t,t)+invgam(t); end
else
try invgam = model.gam(i,1).^-1; catch, invgam = model.gam(1,1).^-1;end
for t=1:model.nb_data, H(t,t) = H(t,t)+invgam; end
end
v = H(model.selector,model.selector)\model.ytrain(model.selector,i);
%eval('v = pcg(H,model.ytrain(model.selector,i), 100*eps,model.nb_data);','v = H\model.ytrain(model.selector, i);');
nu = H(model.selector,model.selector)\ones(nb_data,1);
%eval('nu = pcg(H,ones(model.nb_data,i), 100*eps,model.nb_data);','nu = H\ones(model.nb_data,i);');
s = ones(1,nb_data)*nu(:,1);
model.b(i) = (nu(:,1)'*model.ytrain(model.selector,i))./s;
model.alpha(model.selector,i) = v(:,1)-(nu(:,1)*model.b(i));
end
% Copyright (c) 2010, KULeuven-ESAT-SCD, License & help @ http://www.esat.kuleuven.be/sista/lssvmlab
disp(' This demo illustrates facilities of LS-SVMlab');
disp(' with respect to unsupervised learning.');
disp(' a demo dataset is generated...');
clear yin yang samplesyin samplesyang mema
% initiate variables and construct the data
nb =200;
sig = .20;
% construct data
leng = 1;
for t=1:nb,
yin(t,:) = [2.*sin(t/nb*pi*leng) 2.*cos(.61*t/nb*pi*leng) (t/nb*sig)];
yang(t,:) = [-2.*sin(t/nb*pi*leng) .45-2.*cos(.61*t/nb*pi*leng) (t/nb*sig)];
samplesyin(t,:) = [yin(t,1)+yin(t,3).*randn yin(t,2)+yin(t,3).*randn];
samplesyang(t,:) = [yang(t,1)+yang(t,3).*randn yang(t,2)+yang(t,3).*randn];
end
% plot the data
figure; hold on;
plot(samplesyin(:,1),samplesyin(:,2),'+','Color',[0.6 0.6 0.6]);
plot(samplesyang(:,1),samplesyang(:,2),'+','Color',[0.6 0.6 0.6]);
xlabel('X_1');
ylabel('X_2');
title('Structured dataset');
disp(' (press any key)');
pause
%
% kernel based Principal Component Analysis
%
disp(' ');
disp(' extract the principal eigenvectors in feature space');
disp(' >> nb_pcs=4;'); nb_pcs = 4;
disp(' >> sig2 = .8;'); sig2 = .8;
disp(' >> [lam,U] = kpca([samplesyin;samplesyang],''RBF_kernel'',sig2,[],''eigs'',nb_pcs); ');
[lam,U] = kpca([samplesyin;samplesyang],'RBF_kernel',sig2,[],'eigs',nb_pcs);
disp(' (press any key)');
pause
%
% make a grid over the inputspace
%
disp(' ');
disp(' make a grid over the inputspace:');
disp('>> Xax = -3:0.1:3; Yax = -2.0:0.1:2.5;'); Xax = -3:0.1:3; Yax = -2.0:0.1:2.5;
disp('>> [A,B] = meshgrid(Xax,Yax);'); [A,B] = meshgrid(Xax,Yax);
disp('>> grid = [reshape(A,prod(size(A)),1) reshape(B,1,prod(size(B)))'']; ');三、运行结果



