1 算法过程
(1)随机选取K个簇中心点
(2)通过计算每个样本与每个簇中心点的距离,选择距离最小的簇心,将样本归类到该簇心的簇中
m
i
n
∑
i
=
1
K
∑
x
∈
c
i
d
i
s
t
(
c
i
,
x
)
2
min\sum_{i=1}^K \sum_{x\in c_i}dist(c_i,x)^2
mini=1∑K?x∈ci?∑?dist(ci?,x)2
这里距离可以使用欧几里得距离(Euclidean Distance)、余弦距离(Cosine Distance)、切比雪夫距离(Chebyshew Distance)或曼哈顿距离(Manhattan Distance),计算距离之前需要先对特征值进行标准化。
3、在已经初次分配的簇中,计算该簇中所有向量的均值,作为该的簇中心点
4、重复步骤2和3来进行一定数量的迭代,直到簇中心点在迭代之间变化不大
评价指标
轮廓系数(Silhouette Coefficient),是聚类效果好坏的一种评价方式。 轮廓系数的值是介于 [-1,1] ,越趋近于1代表内聚度和分离度都相对较优。
2 实现
2.1 Python代码
# kmeans算法
# 1. 初始化k个族中心
# 2. 计算每个样本与k个族中心距离,标记样本为距离最短的族
# 3. 重新确定K族中心(族中平均位置)
# 4. 循环2-3,直到前后两次所有族中心距离变化<eps
"""
# 输入:
X: 2d数组, 形如(n_samples, m_features),n_samples表示样本数,m_features表示特征维数
K: int, 超参数,指定族格式
metric:str, 距离类型,默认为欧式距离'Euler',其他暂时为实现
eps: float, 精度(当族中心位置更新变化<eps时停止)
random_state: 随机种子
# 输出:
centers: K族中心向量, 2d数组, 形如(K, m_features)
pred: 1-d数组,长度为n_samples
"""
import numpy as np
import random # 用python的random模块,不用numpy的random
import matplotlib.pyplot as plt
from sklearn.datasets import make_blobs
from scipy.spatial.distance import pdist
from sklearn.preprocessing import StandardScaler
class kmeans: # 创建kmeans类
# 初始化函数
def __init__(self, X=None, K=2, metric='Euler',eps=1e-6, init_centers=None, random_state=None):
self.X = X
self.K = K
self.metric = metric
self.eps = eps
self.centers = init_centers
self.random_state = random_state
# 距离函数
def calc_dist(self, x, c):
dist_list = []
for i in range(c.shape[0]):
v = c[i]
# 欧式距离
if self.metric == 'Euler':
lp = 2
distance = np.power(np.power(x-v, lp), 1/lp).sum()
dist_list.append(distance)
# 切比雪夫距离
elif self.metric == 'Chebyshew':
distance = abs((x)-v).max()
dist_list.append(distance)
# 曼哈顿距离
elif self.metric == 'Manhattan':
distance = sum(abs(x-v))
dist_list.append(distance)
# jaccard距离
elif self.metric == 'Jaccard':
matV = np.vstack([x, v])
distance = pdist(matV, 'jaccard')
dist_list.append(distance)
# 余弦距离
elif self.metric == 'Cosine':
distance = np.dot(x, v) /(np.linalg.norm(x)*np.linalg.norm(v))
dist_list.append(distance)
return dist_list
# 迭代(训练)
def fit(self, X):
# 样本
if X is not None:
self.X = X
# 样本形状
n_samples, n_features = self.X.shape
# 设置随机种子
if self.random_state is not None:
random.seed(self.random_state)
# 初始化聚类中心
if self.centers is None:
# 随机选取簇心(这点可改进,比如kmeans++就是在此改进的)
idx = idx = random.sample(range(n_samples), self.K)
self.centers = X[idx, :]
# 初始样本的族标记-1
pred = np.array([-1]*n_samples)
iter = 0
stop = False # 结束标志
while (not stop):
iter += 1
print(iter)
for i in range(n_samples):
dists = self.calc_dist(X[i, :], self.centers)
pred[i] = np.argmin(dists)
# 重新确定族中心
new_centers = np.zeros((self.K, n_features))
for k in range(self.K):
new_centers[k, :] = X[pred == k, :].mean(axis=0)
# 判断停止条件
delta = abs(new_centers - self.centers)
flg = delta < self.eps
stop = flg.all()
self.centers = new_centers
return pred, self.centers
# 族预测
def predict(self, X):
# 遍历所有样本,划分族
pred = np.array([-1]*n_samples)
for i in range(n_samples):
dists = self.calc_dist(X[i, :], self.centers)
pred[i] = np.argmin(dists)
return pred
if __name__ == "__main__":
# 生成数据
n_samples = 1500
random_state = 170
X, y = make_blobs(n_samples=n_samples, random_state=random_state)
# 调用kmeans
metric_dist = 'Cosine'
model = kmeans(X, K=3, metric=metric_dist, eps=1e-3, random_state=1)
ss = StandardScaler()
X_n = ss.fit_transform(X)
pred, centers = model.fit(X_n)
n_samples, _ = X.shape
# 评价指标:轮廓系数,【-1 1】越接近1 ,聚类效果越好
score = silhouette_score(X, pred)
# 族预测,如果仅是训练数据,直接用fit(X)返回的族划分
# pred = model.predict(X)
plt.scatter(X[:, 0], X[:, 1], c=pred)
plt.title("{}—kmeans".format(metric_dist))
plt.show()
print()
2.2 实验结果
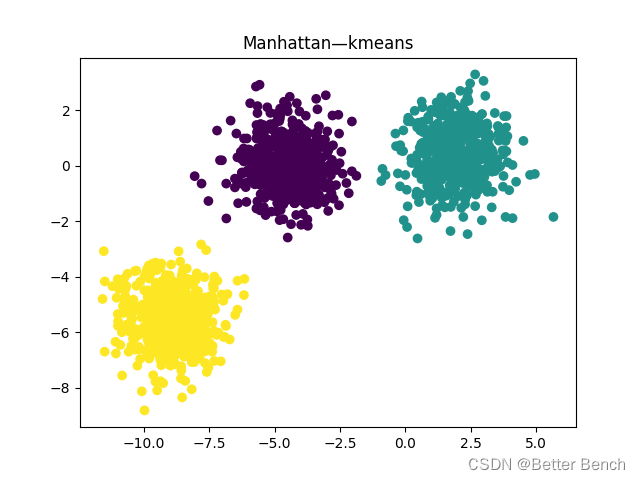
(1)曼哈顿距离
轮廓系数:0.7333423486262539
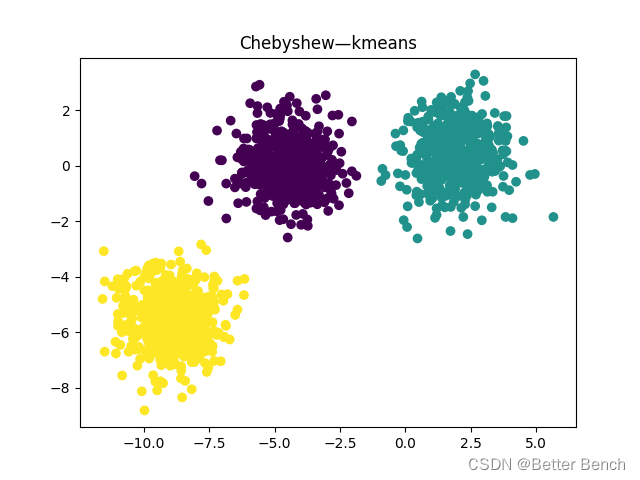
(2)切比雪夫距离
轮廓系数0.7333423486262539
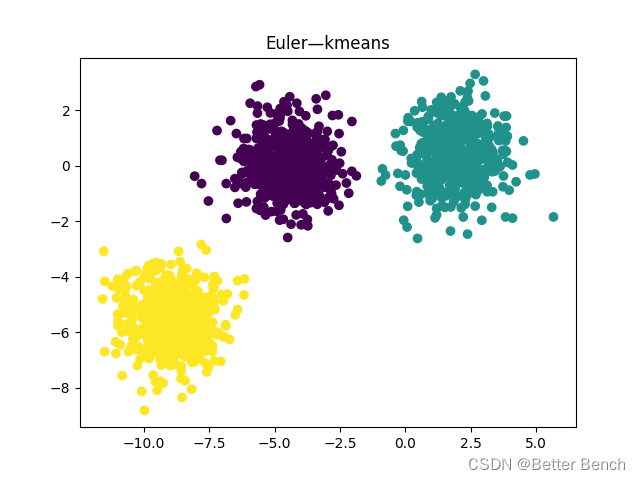
(3)欧式距离
轮廓系数:0.7333423486262539