最近几天实习生和新同事也越来越多了,使用我这台EC2工作的小伙伴也越来越多了。前几天代码运行的到没什么问题,很丝滑很流畅。但从前天开始我的elasticsearch和python脚本是不是的就被killed,一开始我还以为是哪个同事干的,但到了昨天被killed的次数越来越多,我不禁疑惑是谁这么缺德,于是我查看了日志:
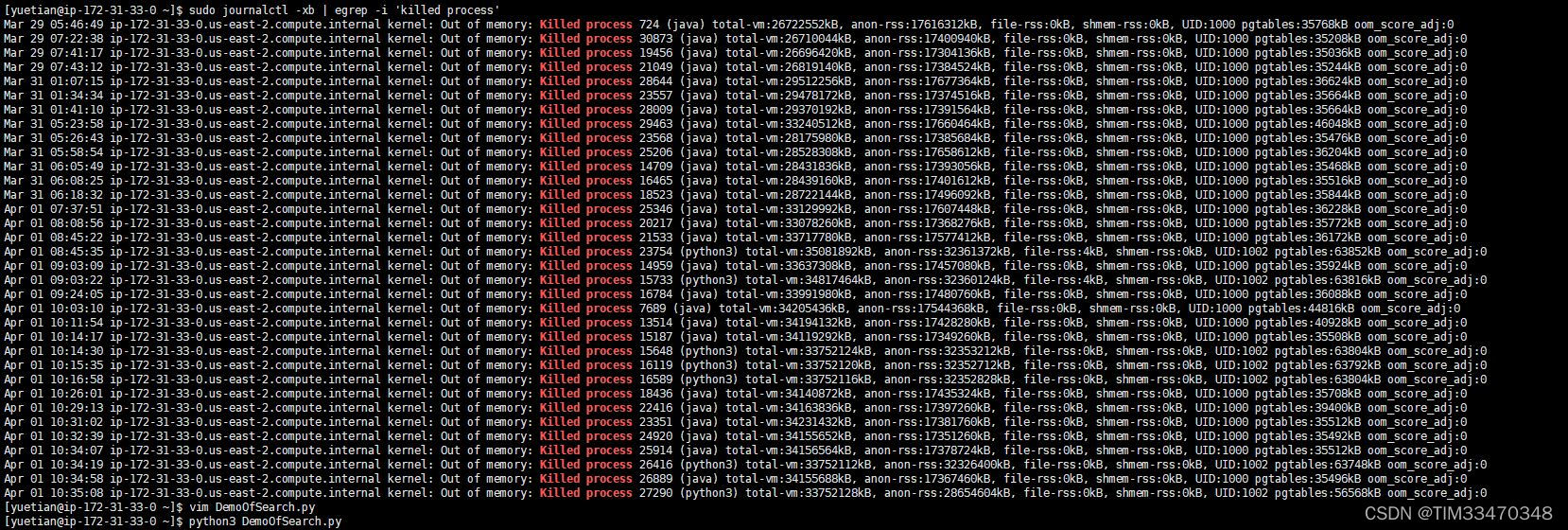
?发现是计算机自己把我的进程给Kill的,原因也写的很清楚:out of memory,也就是爆栈,看来该为这一切背锅的是我自己。
我反思了下爆栈的原因。这是我原来的代码:
from lxml import etree
import pandas as pd
import numpy as np
from elasticsearch import Elasticsearch
# get data from xml file
def get_data(address):
xml = etree.parse(address)
root = xml.getroot()
results = []
datas = root.xpath('//PubmedArticle/MedlineCitation')
date_completed = []
date_revised = []
ISSN = []
DOI = []
ID = []
AbstractList = []
Title = []
Names = []
for data in datas:
# print(data)
if len(data.xpath('./DateCompleted')) != 0:
year = data.xpath('./DateCompleted/Year/text()')[0]
month = data.xpath('./DateCompleted/Month/text()')[0]
day = data.xpath('./DateCompleted/Day/text()')[0]
Date = month+'-'+day+'-'+year
# print(Date)
date_completed.append(Date)
else:
date_completed.append('NULL')
year = data.xpath('./DateRevised/Year/text()')[0]
month = data.xpath('./DateRevised/Month/text()')[0]
day = data.xpath('./DateRevised/Day/text()')[0]
Date = month + '-' + day + '-' + year
date_revised.append(Date)
if len(data.xpath('./Article/Journal/ISSN/text()')) != 0:
issn = data.xpath('./Article/Journal/ISSN/text()')[0]
# print(issn)
ISSN.append(issn)
else:
ISSN.append('NULL')
id = data.xpath('./PMID/text()')[0]
if id != '':
ID.append(id)
else:
ID.append('NULL')
if len(data.xpath('./parent::*/PubmedData/ArticleIdList/ArticleId[@IdType="doi"]/text()')) != 0:
doi = data.xpath('./parent::*/PubmedData/ArticleIdList/ArticleId[@IdType="doi"]/text()')[0]
# print(doi)
DOI.append(doi)
else:
DOI.append('0')
title = data.xpath('./Article/ArticleTitle/text()')
if len(title) != 0:
# print(title)
Title.append(title[0])
else:
Title.append('NULL')
Abstract = data.xpath('./Article/Abstract/AbstractText/text()')
if len(Abstract) == 1:
AbstractList.append(Abstract[0])
# print(Abstract)
elif len(Abstract) == 0:
AbstractList.append('NULL')
else:
length = len(Abstract)
# print(length)
AbstractString = ''
for tmp in Abstract:
AbstractString.join(tmp)
# Abstract = data.xpath('./Article/Abstract/AbstractText/text()')[0]
# Abstract = data.xpath('./Article/Abstract/AbstractText[@Label="BACKGROUND"]/text()')[0]
AbstractList.append(AbstractString)
# print(AbstractString)
authorLastName = data.xpath('./Article/AuthorList/Author/LastName/text()')
authorForeName = data.xpath('./Article/AuthorList/Author/ForeName/text()')
authorName = []
if len(authorForeName) != 0 and len(authorLastName) != 0 and len(authorForeName) == len(authorLastName):
for i in range(len(authorLastName)):
authorName.append(authorForeName[i] + ' ' + authorLastName[i] + ',')
# print(authorName)
Names.append("".join(authorName))
else:
Names.append("NULL")
final = pd.DataFrame(np.array([date_completed, date_revised, ISSN, DOI, AbstractList, Title, ID, Names]).T,
columns=['dateCompleted', 'DateRevised', 'ISSN', 'DOI', 'AbstractList', 'Title',
'PMID', 'Authors'])
# final.to_csv('c:\\result.csv')
return final
# PUT data into Elasticsearch
def write_data(data):
es = Elasticsearch(hosts=['localhost:9200'])
index_name = "pubmed-paper-index"
length = len(data['PMID'])
for i in range(length):
jsonData = {
"title": data.loc[i]['Title'],
"Authors": data.loc[i]['Authors'],
"abstract": data.loc[i]['AbstractList'],
"date": {
"completed_date": data.loc[i]['dateCompleted'],
"revised_date": data.loc[i]['DateRevised']
},
"journal_issn": data.loc[i]['ISSN'],
"journal_doi": data.loc[i]['DOI'],
"article_id": data.loc[i]['PMID']
}
print(jsonData)
es.index(index=index_name, body=jsonData)
if __name__ == '__main__':
# List of pending documents
address = [
'pubmed22n0001.xml',
'pubmed22n0002.xml',
'pubmed22n0003.xml',
]
# process the file
for add in address:
data = get_data(add)
write_data(data)可以看到,我get_data函数中先从xml文件里读出所有数据放入pandas的dataframe中,然后再用write_data函数逐个写入elasticsearch中,这样dataframe数据结构就一下子存放了一个xml文件的所有需要的内容,这是一个很庞大的文件,加之小伙伴们也在用EC2,所以就爆栈了。
那解决办法也是不言而喻的,就是在读xml文件时,每读一条数据就把这条数据写入elasticsearch中,这样就避免了大量占用内寸的问题。改进的代码如下:
import logging
from lxml import etree
from elasticsearch import Elasticsearch
# get data from xml file
def get_data(address):
xml = etree.parse(address)
root = xml.getroot()
datas = root.xpath('//PubmedArticle/MedlineCitation')
es = Elasticsearch(hosts=['http://localhost:9200'])
for data in datas:
# print(data)
if len(data.xpath('./DateCompleted')) != 0:
year = data.xpath('./DateCompleted/Year/text()')[0]
month = data.xpath('./DateCompleted/Month/text()')[0]
day = data.xpath('./DateCompleted/Day/text()')[0]
Date = month+'-'+day+'-'+year
# print(Date)
date_completed = Date
else:
date_completed = 'NULL'
year = data.xpath('./DateRevised/Year/text()')[0]
month = data.xpath('./DateRevised/Month/text()')[0]
day = data.xpath('./DateRevised/Day/text()')[0]
Date = month + '-' + day + '-' + year
date_revised = Date
if len(data.xpath('./Article/Journal/ISSN/text()')) != 0:
issn = data.xpath('./Article/Journal/ISSN/text()')[0]
# print(issn)
ISSN = issn
else:
ISSN = 'NULL'
id = data.xpath('./PMID/text()')[0]
if id != '':
ID = id
else:
ID = 'NULL'
if len(data.xpath('./parent::*/PubmedData/ArticleIdList/ArticleId[@IdType="doi"]/text()')) != 0:
doi = data.xpath('./parent::*/PubmedData/ArticleIdList/ArticleId[@IdType="doi"]/text()')[0]
# print(doi)
DOI = doi
else:
DOI = '0'
title = data.xpath('./Article/ArticleTitle/text()')
if len(title) != 0:
# print(title)
Title = title[0]
else:
Title = 'NULL'
Abstract = data.xpath('./Article/Abstract/AbstractText/text()')
if len(Abstract) == 1:
AbstractList = Abstract[0]
# print(Abstract)
elif len(Abstract) == 0:
AbstractList = 'NULL'
else:
length = len(Abstract)
# print(length)
AbstractString = ''
for tmp in Abstract:
AbstractString.join(tmp)
# Abstract = data.xpath('./Article/Abstract/AbstractText/text()')[0]
# Abstract = data.xpath('./Article/Abstract/AbstractText[@Label="BACKGROUND"]/text()')[0]
AbstractList = AbstractString
# print(AbstractString)
authorLastName = data.xpath('./Article/AuthorList/Author/LastName/text()')
authorForeName = data.xpath('./Article/AuthorList/Author/ForeName/text()')
authorName = []
if len(authorForeName) != 0 and len(authorLastName) != 0 and len(authorForeName) == len(authorLastName):
for i in range(len(authorLastName)):
authorName.append(authorForeName[i] + ' ' + authorLastName[i] + ',')
# print(authorName)
Names = "".join(authorName)
else:
Names = "NULL"
jsonData = {
"title": Title,
"Authors": Names,
"abstract": AbstractList,
"date": {
"completed_date": date_completed,
"revised_date": date_revised,
},
"journal_issn": ISSN,
"journal_doi": DOI,
"article_id": ID
}
index_name = "pubmed-paper-index"
es.index(index=index_name, body=jsonData)
if __name__ == '__main__':
# List of pending documents
address = []
path1 = "pubmed22n"
path2 = ".xml.gz"
path = ''
for i in range(715, 1115):
if i >0 and i <= 9:
path = path1 + '000' + str(i) +path2
address.append(path)
elif i >= 10 and i<= 99:
path = path1 + '00' + str(i) + path2
address.append(path)
elif i >= 100 and i <= 999:
path = path1 + '0' + str(i) + path2
address.append(path)
else:
path = path1 + str(i) + path2
address.append(path)
# process the file
for add in address:
get_data(add)
logging.basicConfig()
logger = logging.getLogger(__name__)
logger.setLevel(logging.INFO)
logger.info(add + f"finished!")
可以看到,log的显示和之前仍然保持一致,每写完一个文件都会报告一次进度。
END